Generating and Visualizing Topic Models with Tethne and MALLET¶
This tutorial was developed for the course Introduction to Digital & Computational Methods in the Humanities (HPS), created and taught by Julia Damerow and Erick Peirson.
Tethne provides a variety of methods for working with text corpora and the output of modeling tools like MALLET. This tutorial focuses on parsing, modeling, and visualizing a Latent Dirichlet Allocation topic model, using data from the JSTOR Data-for-Research portal.
In this tutorial, we will use Tethne to prepare a JSTOR DfR corpus for topic modeling in MALLET, and then use the results to generate a semantic network like the one shown below.
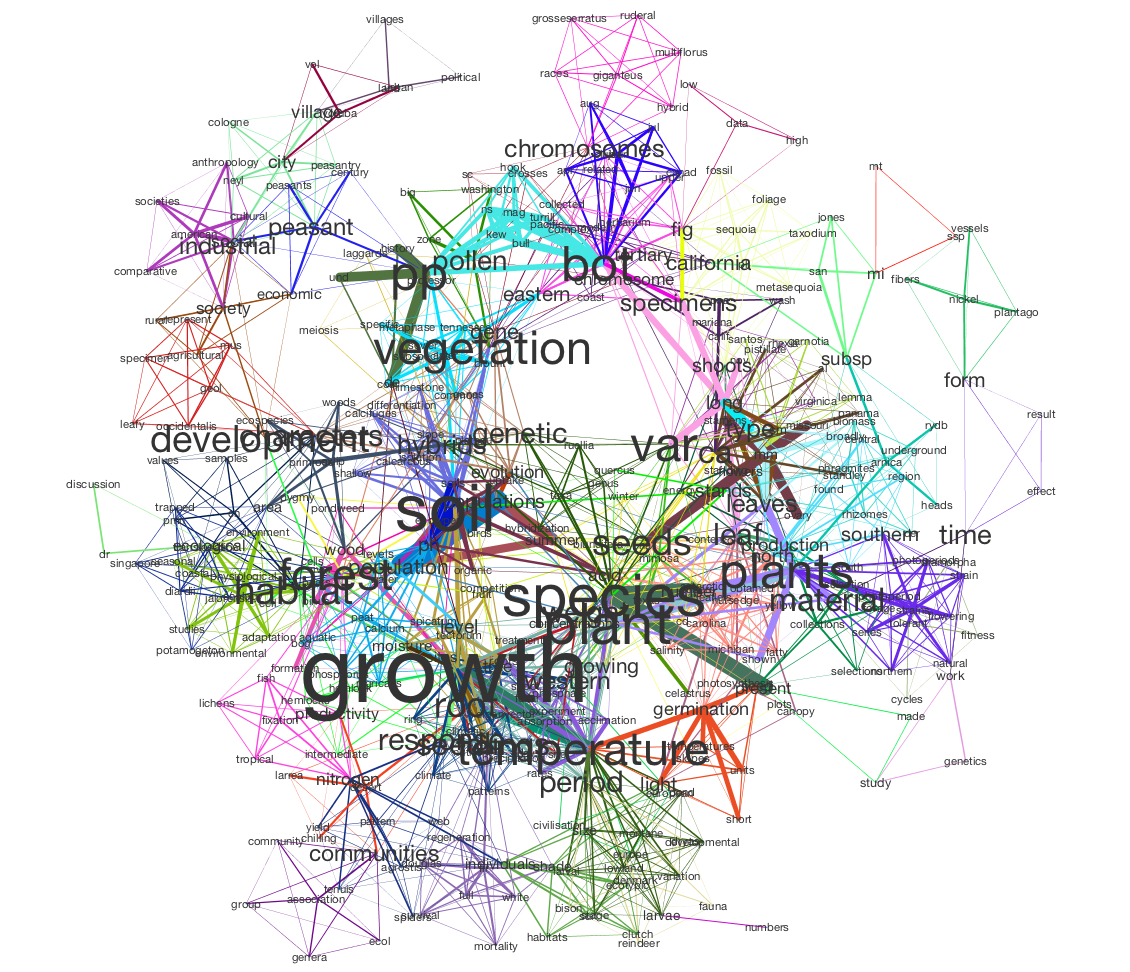
In this visualization, words are connected if they are associated with the same topic; the heavier the edge, the more strongly those words are associated with that topic. Each topic is represented by a different color. The size of each word indicates the structural importance (betweenness centrality) of that word in the semantic network.
As of v0.4, corpus-oriented methods have not yet been implemented in the Tethne command-line interface or GUI.
This tutorial assumes that you already have a basic familiarity with Cytoscape.
Before You Start¶
You’ll need some data. See JSTOR Data-for-Research for instructions on retrieving data. Note that Tethne currently only supports XML output from JSTOR. Be sure to get some wordcounts so that you’ll have some data for modeling.
Be sure that you have the latest release of Tethne. See Installation.
You should also download and install MALLET. It’s also not a bad idea to check out this tutorial for topic modeling with MALLET.
Loading JSTOR DfR¶
tethne.readers.dfr provides two mechanisms for loadings data from JSTOR DfR:
- dfr.read() loads bibliographic records from the
citations.XML file accompanying the dataset. This isn’t particularly necessary for the purpose of this exercise, but is worth knowing about.
- dfr.ngrams() loads N-grams (including unigrams/wordcounts)
from the dataset. We’ll use these as the raw data for topic modeling.
Assuming that you unzipped your JSTOR DfR dataset to /Users/erickpeirson/JStor DfR Datasets/2013.5.3.cHrmED8A, you can use something like the following to load wordcounts from your dataset:
>>> import tethne.readers as rd
>>> datapath = '/Users/erickpeirson/JStor DfR Datasets/2013.5.3.cHrmED8A'
>>> wordcounts = rd.dfr.ngrams(datapath, N='uni')
wordcounts should now contain a dictionary mapping each paper (by DOI) to a list of (word, frequency) tuples. For example:
>>> wordcounts.keys()[0:3]
['10.2307/1293500', '10.2307/1936479', '10.2307/2433815']
>>> wordcounts['10.2307/1293500'][0:6]
[('the', 49), ('of', 49), ('in', 33), ('and', 29), ('a', 21), ('to', 21)]
Generating Documents¶
One of the most straight-forward ways to load documents into MALLET for topic modeling is to pass it a plain-text file containing the full text of each document on its own line. Since JSTOR DfR data consist only of term frequencies for each document, we’ll need to reconstruct each document. Since word order doesn’t matter in LDA topic modeling, we can write a document by simply repeating each term by its corresponding frequency. For example, these term frequencies...
[('microbiology', 7), ('with', 7), ('are', 7), ('have', 7), ('be', 7),
('is', 6), ('issue', 6), ('training', 6), ('g', 6), ('bioscience', 6)]
...would result in the document...
'microbiology microbiology microbiology microbiology microbiology
microbiology microbiology with with with with with with with are are
are are are are are have have have have have have have be be be be be
be be is is is is is is issue issue issue issue issue issue training
training training training training training g g g g g g bioscience
bioscience bioscience bioscience bioscience bioscience'
We can use tethne.writers.corpora.to_documents() to generate such a corpus.
>>> import tethne.writers as wr
>>> wr.corpora.to_documents('./mycorpus', wordcounts)
This generates a text file called mycorpus_docs.txt containing all of our documents, and a file called mycorpus_meta.csv that maps each row in the corpus to a DOI.
Topic Modeling in MALLET¶
For details about LDA modeling in MALLET, consult the MALLET website as well as this tutorial.
First, tell MALLET to load the corpus that Tethne generated for you. Following the example on the MALLET website, use something like:
$ bin/mallet import-file --input /Users/erickpeirson/mycorpus_docs.txt \
> --output mytopic-input.mallet --keep-sequence --remove-stopwords
When you train your model, you’ll want to specify a few output options so that Tethne will have something to work with later: --output-doc-topics, --word-topic-counts-file, and --output-topic-keys:
$ bin/mallet train-topics --input mytopic-input.mallet --num-topics 100 \
> --output-doc-topics /Users/erickpeirson/doc_top \
> --word-topic-counts-file /Users/erickpeirson/word_top \
> --output-topic-keys /Users/erickpeirson/topic_keys
Modeling should commence, and run for a few minutes (or longer, depending on the size of your corpus and the number of iterations). Note that we chose 100 topics in the example above.
<1000> LL/token: -8.62952
Total time: 1 minutes 12 seconds
$
Parsing MALLET Output¶
Tethne can read MALLET output using the methods in tethne.readers.mallet:
- mallet.load() parses MALLET output, and generates a LDAModel object that can be used for subsequent analysis and visualization.
- mallet.read() behaves like the read method in any other tethne.readers sub-module, and generates a list of Paper objects with vectors from the LDAModel attached.
We’ll start by passing mallet.load() paths to the MALLET output files from the previous step:
>>> import tethne.readers as rd
>>> td = '/Users/erickpeirson/doc_top'
>>> tw = '/Users/erickpeirson/word_top'
>>> tk = '/Users/erickpeirson/topic_keys'
>>> m = '/Users/erickpeirson/mycorpus_meta.csv'
>>> Z = 100 # Number of topics
>>> model = rd.mallet.load(td, tw, tk, Z, m)
We can also load up corresponding Paper objects using the same arguments:
>>> papers = rd.mallet.read(td, tw, tk, Z, m)
Semantic Network¶
In LDA, topics are clusters of terms that co-occur in documents. We can interpret an LDA topic model as a network of terms linked by their participation in particular topics. In Tethne, we call this a topic-coupling network.
Build the Network¶
We can generate the topic-coupling network using tethne.networks.terms.topic_coupling():
>>> import tethne.networks as nt
>>> g = nt.terms.topic_coupling(model, threshold=0.015)
The threshold argument tells Tethne the minimum P(W|T) to consider a topic (T) to contain a given word (W). In this example, the threshold was chosen post-hoc by adjusting its value and eye-balling the resultant network for coherence.
We can then write this graph to a GraphML file for visualization:
>>> import tethne.writers as wr
>>> wr.graph.to_graphml(g, './mymodel_tc.graphml')
Visualization¶
In Cytoscape, import your GraphML network by selecting File > Import > Network > From file... and choosing the file mymodel_tc.graphml from the previous step.
Edge weight¶
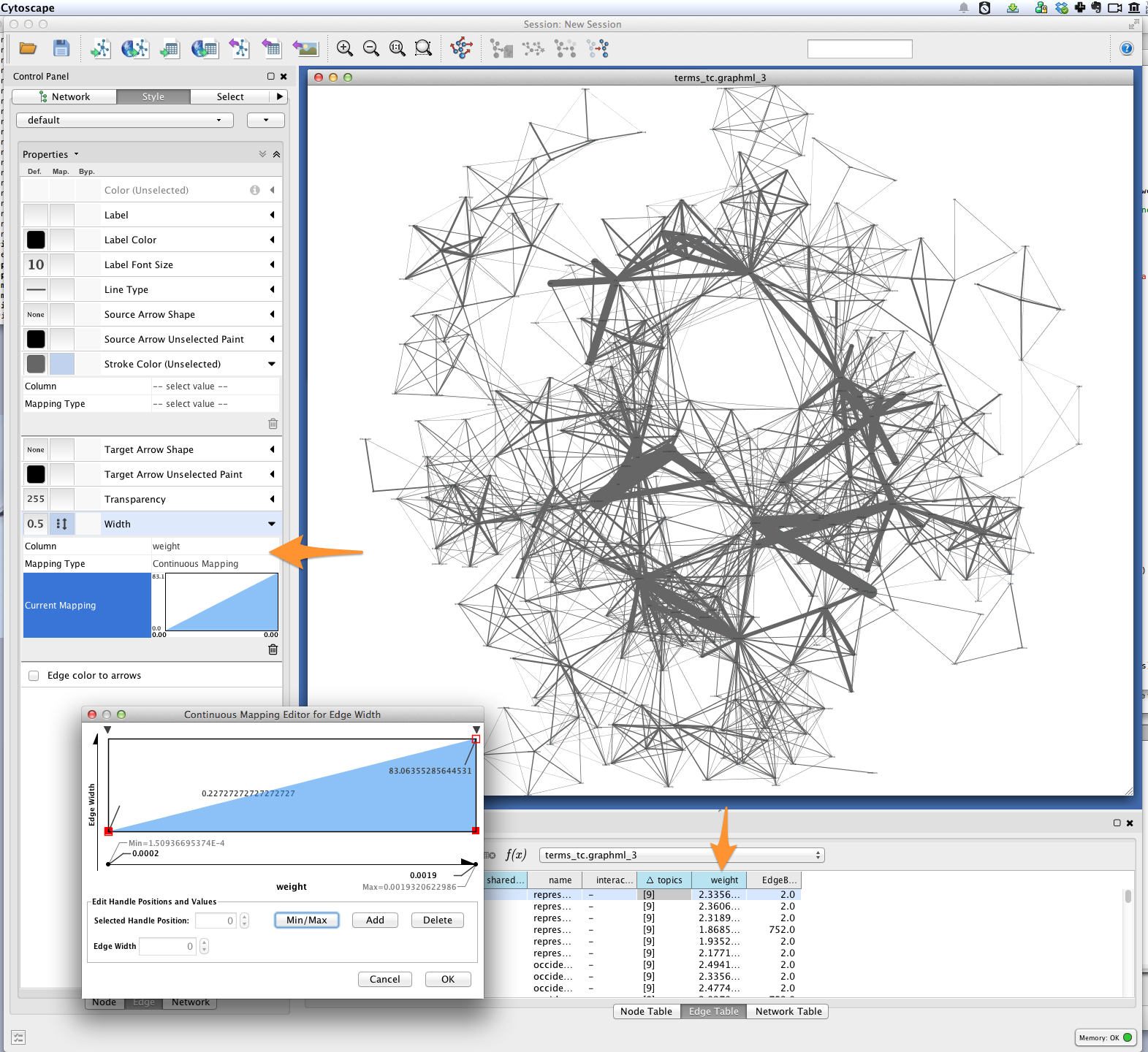
Tethne included joint average P(W|T) for each pair of terms in the graph as the edge attribute weight. You can use this value to improve the layout of your network. Try selecting Layout > Edge-weighted Spring Embedded > weight.
You can also use a continuous mapper to represent edge weights visually. Create a new visual mapping (in the VizMapper tab in Cytoscape < 3.1, Style in >= 3.1) for edge width.
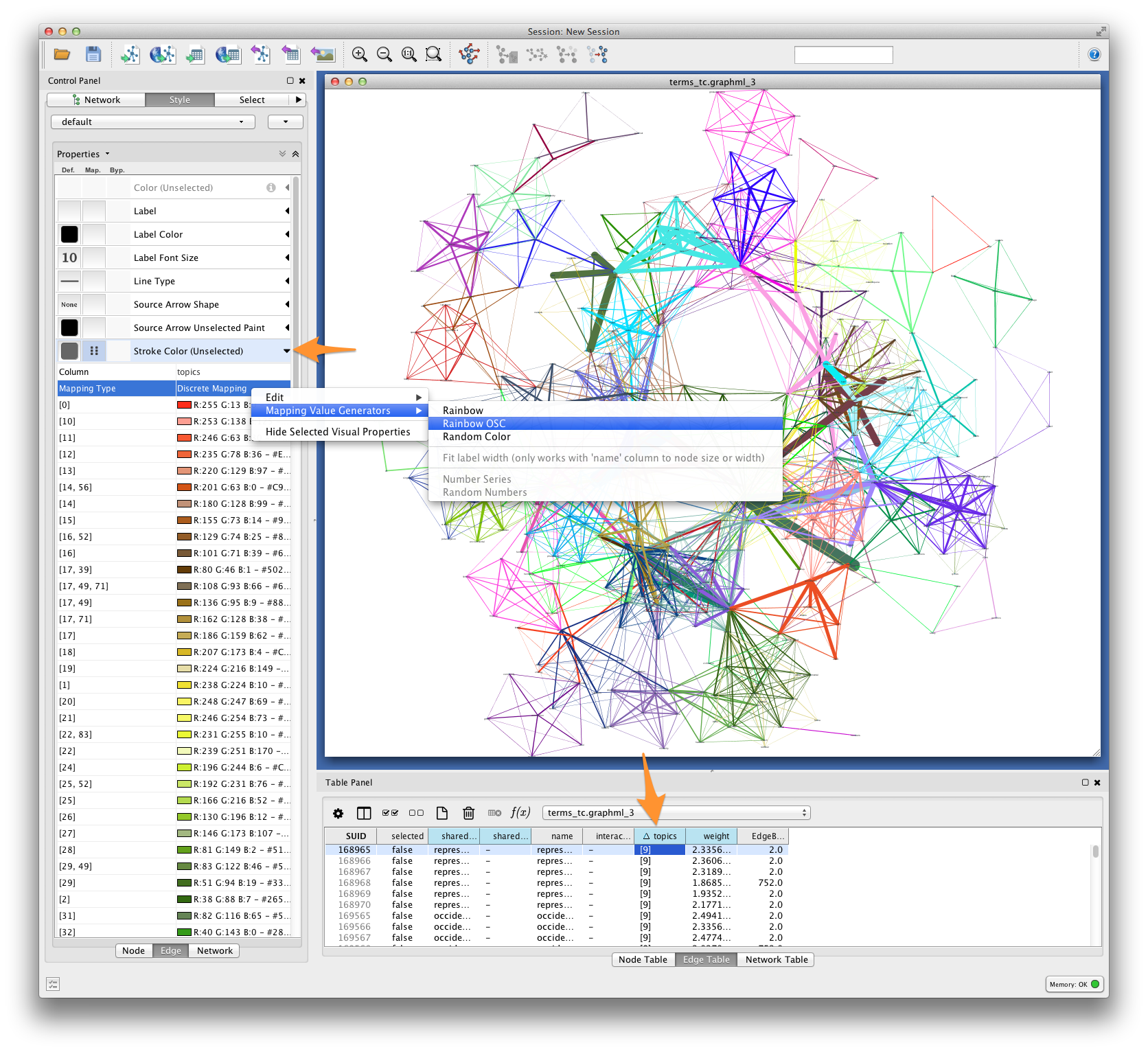
Edge color¶
For each pair of terms, Tethne records shared topics in the edge attribute topics. Coloring edges by shared topic will give a visual impression of the “parts” of your semantic network. Create a discrete mapping for edge stroke color, and then right-click on the mapping to choose a color palette from the Mapping Value Generators.
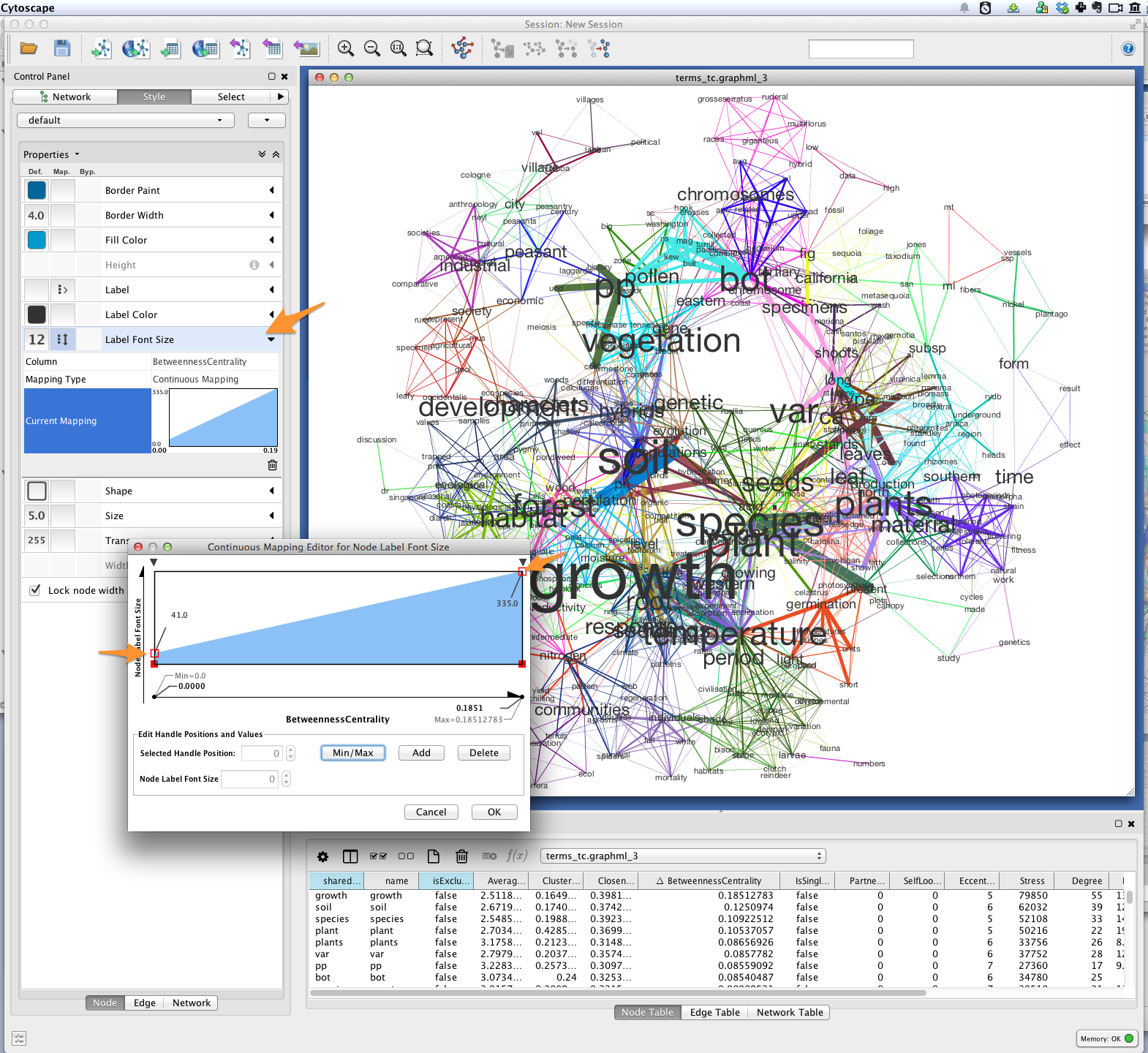
Font-size¶
Finally, you’ll want to see the words represented by each of the nodes in your network. You might be interested in which terms are most responsible for bridging the various topics in your model. This “bridging” role is best captured using betweenness centrality, which is a measure of the structural importance of a given node. Nodes that connect otherwise poorly-connected regions of the network (e.g. clusters of words in a semantic network) have high betweenness-centrality.
Use Cytoscape’s NetworkAnalyzer to generate centrality values for each node: select Tools > NetworkAnalyzer > Network Analysis > Analyze Network. Once analysis is complete, Cytoscape should automatically add a BetweennessCentrality node attribute to the graph.
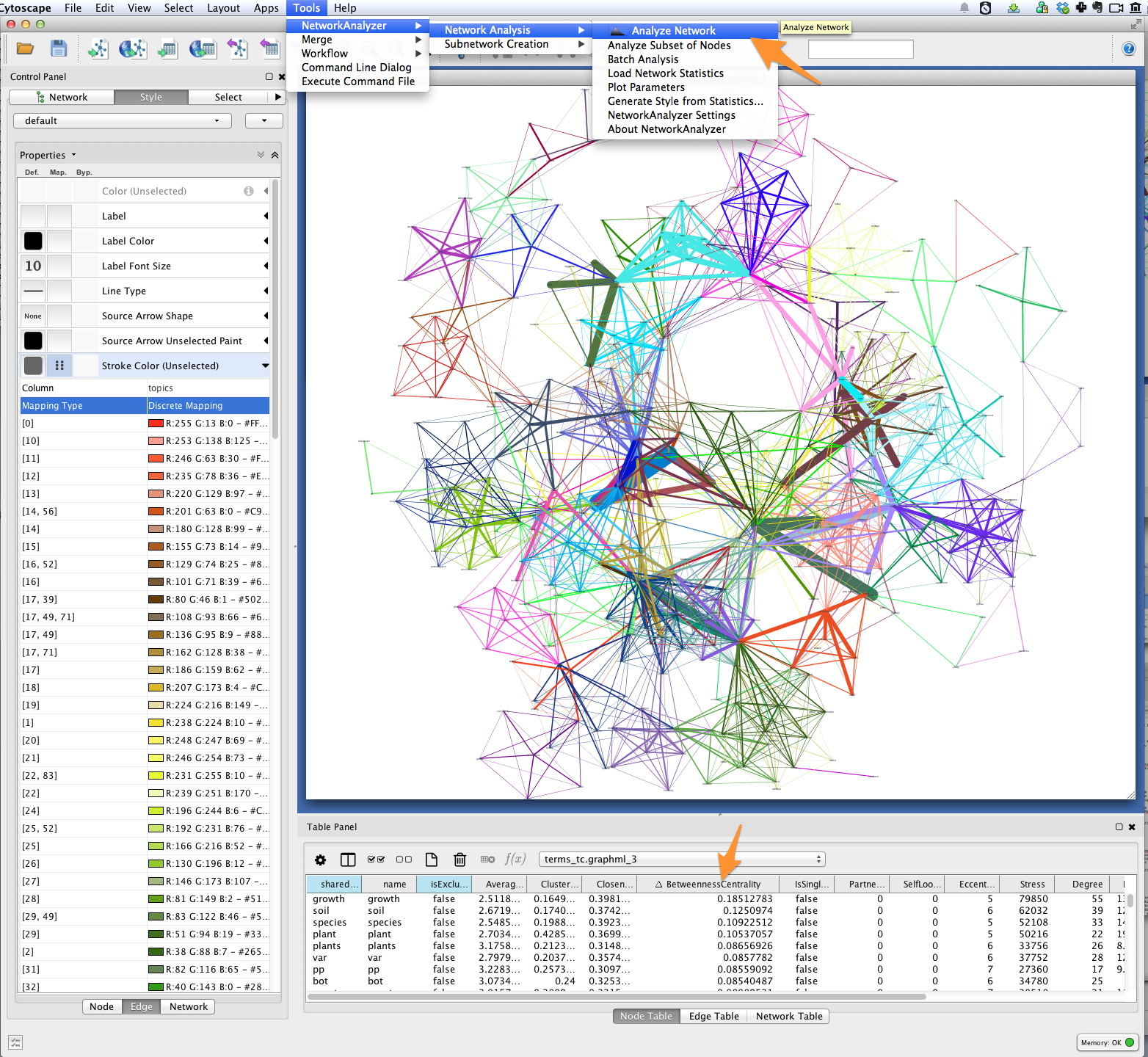
Next, create a continuous mapping for Label Font Size based on BetweennessCentrality. More central words should appear larger. In the figure below, label font size ranges from around 40 to just over 300 pt.
Export¶
Export the finished visualization by selecting File > Export > Network View as Graphics....
Wrapping up, Looking forward¶
To generate a network of papers connected by topics-in-common, try the networks.papers.topic_coupling() method.
Since Tethne is still under active development, methods for working with topic modeling and other corpus-analysis techniques are being added all the time, and existing functions will likely change as we find ways to streamline workflows. This tutorial will be updated and extended as development proceeds.