tethne Package¶
tethne Package¶
Tethne is a package for analyzing citation data from the Web of Science. Modules within Tethne can generate a variety of networks, such as bibliographic coupling, citation, author-paper, and co-author networks, using networkx.
| tethne.analyze | The tethne.analyze sub-package provides additional analysis methods not |
| tethne.builders | Classes for building a GraphCollection . |
| tethne.data | Classes for handling bibliographic data. |
| tethne.matrices | Methods for generating matrices from Paper objects and other data. |
| tethne.networks | Methods for building networks from bibliographic data. |
| tethne.readers | Methods for parsing bibliographic datasets. |
| tethne.services | Modules for interacting with external web services. |
| tethne.utilities | Helper functions for tethne.networks . |
| tethne.writers | Export networks to structured and unstructured formats, for visualization. |
__main__ Module¶
Provides the Tethne command-line interface.
See Quickstart (Command-line) and Command-line Options for an introduction to the CLI.
builders Module¶
Classes for building a GraphCollection .
| builder(D) | Base class for builders. |
| authorCollectionBuilder(D) | Builds a GraphCollection with method in |
| paperCollectionBuilder(D) | Builds a GraphCollection with method in |
Bases: tethne.builders.builder
Builds a GraphCollection with method in tethne.networks.authors from a DataCollection .
Methods
build(graph_axis, graph_type, **kwargs) Generates graphs for each slice along graph_axis in Generates graphs for each slice along graph_axis in DataCollection D.
Other axes in D are treated as attributes.
Usage
>>> import tethne.readers as rd >>> data = rd.wos.read("/Path/to/wos/data.txt") >>> from tethne.data import DataCollection >>> D = DataCollection(data) # Indexed by wosid, by default. >>> D.slice('date', 'time_window', window_size=4) >>> from tethne.builders import authorCollectionBuilder >>> builder = authorCollectionBuilder(D) >>> C = builder.build('date', 'coauthors') >>> C <tethne.data.GraphCollection at 0x104ed3550>
- class tethne.builders.paperCollectionBuilder(D)[source]¶
Bases: tethne.builders.builder
Builds a GraphCollection with method in tethne.networks.papers from a DataCollection .
Methods
build(graph_axis, graph_type, **kwargs) Generates graphs for each slice along graph_axis in - build(graph_axis, graph_type, **kwargs)[source]¶
Generates graphs for each slice along graph_axis in DataCollection D.
Other axes in D are treated as attributes.
Usage
>>> import tethne.readers as rd >>> data = rd.wos.read("/Path/to/wos/data.txt") >>> from tethne.data import DataCollection >>> D = DataCollection(data) # Indexed by wosid, by default. >>> D.slice('date', 'time_window', window_size=4) >>> from tethne.builders import paperCollectionBuilder >>> builder = paperCollectionBuilder(D) >>> C = builder.build('date', 'bibliographic_coupling', threshold=2) >>> C <tethne.data.GraphCollection at 0x104ed3550>
data Module¶
Classes for handling bibliographic data.
| Paper() | Base class for Papers. |
| DataCollection(data[, index_by]) | A DataCollection organizes Papers for analysis. |
| GraphCollection() | Collection of NetworkX nx.classes.graph.Graph objects, |
| LDAModel(doc_topic, top_word, top_keys, ...) | Organizes parsed output from MALLET’s LDA modeling algorithm. |
- class tethne.data.DataCollection(data, index_by='wosid')[source]¶
Bases: object
A DataCollection organizes Papers for analysis.
The DataCollection is initialized with some data, which is indexed by a key in Paper (default is wosid). The DataCollection can then be sliced ( DataCollection.slice() ) by other keys in Paper .
Usage
>>> import tethne.readers as rd >>> data = rd.wos.read("/Path/to/wos/data.txt") >>> data += rd.wos.read("/Path/to/wos/data2.txt") # Two accessions. >>> from tethne.data import DataCollection >>> D = DataCollection(data) # Indexed by wosid, by default. >>> D.slice('date', 'time_window', window_size=4) >>> D.slice('accession') >>> D <tethne.data.DataCollection at 0x10af0ef50>
Methods
N_axes() Returns the number of slice axes for this DataCollection . distribution() Returns a Numpy array describing the number of Paper distribution_2d(x_axis, y_axis) Returns a Numpy array describing the number of Paper get_axes() Returns a list of all slice axes for this DataCollection . get_by(key_indices[, papers]) Given a set of (key, index) tuples, return the corresponding subset of get_slice(key, index[, papers]) Yields a specific slice. get_slices(key[, papers]) Yields slices for key. indices() Yields a list of indices of all papers in this DataCollection papers() Yield the complete set of Paper instances in this slice(key[, method]) Slices data by key, using method (if applicable). - N_axes()[source]¶
Returns the number of slice axes for this DataCollection .
- distribution()[source]¶
Returns a Numpy array describing the number of Paper associated with each slice-coordinate.
WARNING: expensive for a DataCollection with many axes or long axes. Consider using distribution_2d() .
Returns : dist : Numpy array
An N-dimensional array. Axes are given by DataCollection.get_axes() and values are the number of Paper at that slice-coordinate.
Raises : RuntimeError : DataCollection has not been sliced.
- distribution_2d(x_axis, y_axis)[source]¶
Returns a Numpy array describing the number of Paper associated with each slice-coordinate, for x and y axes spcified.
Returns : dist : Numpy array
A 2-dimensional array. Values are the number of Paper at that slice-coordinate.
Raises : RuntimeError : DataCollection has not been sliced.
KeyError: Invalid slice axes for this DataCollection. :
- get_axes()[source]¶
Returns a list of all slice axes for this DataCollection .
- get_by(key_indices, papers=False)[source]¶
Given a set of (key, index) tuples, return the corresponding subset of Paper indices (or Paper instances themselves, if papers is True).
Parameters : key_indices : list
A list of (key, index) tuples.
Returns : plist : list
A list of paper indices, or Paper instances.
Raises : RuntimeError : DataCollection has not been sliced.
- get_slice(key, index, papers=False)[source]¶
Yields a specific slice.
Parameters : key : str
Key from Paper that has previously been used to slice data in this DataCollection .
index : str or int
Slice index for key (e.g. 1999 for ‘date’).
Returns : slice : list
List of paper indices in this DataCollection , or (if papers is True) a list of Paper instances.
Raises : RuntimeError : DataCollection has not been sliced.
KeyError : Data has not been sliced by [key]
KeyError : [index] not a valid index for [key]
- get_slices(key, papers=False)[source]¶
Yields slices for key.
Parameters : key : str
Key from Paper that has previously been used to slice data in this DataCollection .
Returns : slices : dict
Keys are slice indices. If papers is True, values are lists of Paper instances; otherwise returns paper indices (e.g. ‘wosid’).
Raises : RuntimeError : DataCollection has not been sliced.
KeyError : Data has not been sliced by [key]
- indices()[source]¶
Yields a list of indices of all papers in this DataCollection
Returns : list :
List of indices.
- papers()[source]¶
Yield the complete set of Paper instances in this DataCollection .
Returns : papers : list
A list of Paper
- slice(key, method=None, **kwargs)[source]¶
Slices data by key, using method (if applicable).
Methods available for slicing a DataCollection:
Method Description Key kwargs time_window Slices data using a sliding time-window. Dataslices are indexed by the start of the time-window. date window_size step_size time_period Slices data into time periods of equal length. Dataslices are indexed by the start of the time period. date window_size The main difference between the sliding time-window (time_window) and the time-period (time_period) slicing methods are whether the resulting periods can overlap. Whereas time-period slicing divides data into subsets by sequential non-overlapping time periods, subsets generated by time-window slicing can overlap.

Time-period slicing, with a window-size of 4 years.
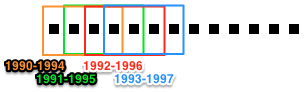
Time-window slicing, with a window-size of 4 years and a step-size of 1 year.
Avilable kwargs:
Argument Type Description window_size int Size of time-window or period, in years (default = 1). step_size int Amount to advance time-window or period in each step (ignored for time_period). cumulative bool If True, the data from each successive slice includes the data from all preceding slices. Only applies if key is ‘date’ (default = False). Parameters : key : str
key in Paper by which to slice data.
method : str (optional)
Dictates how data should be sliced. See table for available methods. If key is ‘date’, default method is time_period with window_size and step_size of 1.
kwargs : kwargs
See methods table, above.
- class tethne.data.GraphCollection[source]¶
Bases: object
Collection of NetworkX nx.classes.graph.Graph objects, organized by some index (e.g. time).
A GraphCollection can be generated using classes in the tethne.builders module. See Generating a GraphCollection from a DataCollection for details.
Methods
compose() Returns the simple union of all Graph in the edges([overwrite]) Return complete set of edges for this GraphCollection . load(filepath) Loads a pickled (serialized) GraphCollection from filepath. nodes([overwrite]) Return complete set of nodes for this GraphCollection . save(filepath) Pickles (serializes) the GraphCollection . - compose()[source]¶
Returns the simple union of all Graph in the GraphCollection .
Returns : composed : Graph
Simple union of all Graph in the GraphCollection .
Notes
Node or edge attributes that vary over slices should be ignored.
- edges(overwrite=False)[source]¶
Return complete set of edges for this GraphCollection .
If this method has been called previously for this GraphCollection then will not recompute unless overwrite = True.
Parameters : overwrite : bool
If True, will generate new node list, even if one already exists.
Returns : edges : list
List (complete set) of edges for this GraphCollection .
- load(filepath)[source]¶
Loads a pickled (serialized) GraphCollection from filepath.
Parameters : filepath : string
Full path to pickled GraphCollection .
Raises : UnpicklingError : Raised when there is some issue in unpickling.
IOError : File does not exist, or cannot be read.
- nodes(overwrite=False)[source]¶
Return complete set of nodes for this GraphCollection .
If this method has been called previously for this GraphCollection then will not recompute unless overwrite = True.
Parameters : overwrite : bool
If True, will generate new node list, even if one already exists.
Returns : nodes : list
List (complete set) of node identifiers for this GraphCollection .
- save(filepath)[source]¶
Pickles (serializes) the GraphCollection .
Parameters : filepath : :
Full path of output file.
Raises : PicklingError : Raised when unpicklable objects are Pickled.
IOError : File does not exist, or cannot be opened.
- class tethne.data.LDAModel(doc_topic, top_word, top_keys, metadata, vocabulary)[source]¶
Bases: object
Organizes parsed output from MALLET’s LDA modeling algorithm.
Used by readers.mallet.
Methods
docs_in_topic(z[, topD]) Returns a list of the topD documents most representative of topic z. topics_in_doc(d[, topZ]) Returns a list of the topZ most prominent topics in a document. - docs_in_topic(z, topD=None)[source]¶
Returns a list of the topD documents most representative of topic z.
Parameters : z : int
A topic index.
topD : int or float
Number of prominent topics to return (int), or threshold (float).
Returns : documents : list
List of (document, proportion) tuples.
- topics_in_doc(d, topZ=None)[source]¶
Returns a list of the topZ most prominent topics in a document.
Parameters : d : str or int
An identifier from a Paper key.
topZ : int or float
Number of prominent topics to return (int), or threshold (float).
Returns : topics : list
List of (topic, proportion) tuples.
- class tethne.data.Paper[source]¶
Bases: object
Base class for Papers.
Behaves just like a dict, but enforces a limited vocabulary of keys, and specific data types.
The following fields (and corresponding data types) are allowed:
Field Type Description aulast list Authors’ last name, as a list. auinit list Authors’ first initial as a list. institution dict Institutions with which the authors are affiliated. atitle str Article title. jtitle str Journal title or abbreviated title. volume str Journal volume number. issue str Journal issue number. spage str Starting page of article in journal. epage str Ending page of article in journal. date int Article date of publication. country dict Author-Country mapping. citations list A list of Paper instances. ayjid str First author’s name (last fi), pubdate, and journal. doi str Digital Object Identifier. pmid str PubMed ID. wosid str Web of Science UT fieldtag value. accession str Identifier for data conversion accession. None values are also allowed for all fields.
Methods
authors() Returns a list of author names (FI LAST). iteritems() Returns an iterator for the Paper‘s metadata fields keys() Returns the keys of the Paper‘s metadata fields. values() Returns the values of the Paper‘s metadata fields. Returns a list of author names (FI LAST).
workflow Module¶
Methods for network analysis.
- tethne.workflow.closeness_introgression(papers, node, window_size, normalize=False)[source]¶
Analyzes the global closeness centrality of a node over time.
Parameters : papers : list
A list of Paper instances.
node : any
Handle of the node to analyze.
window_size : int
Size of time-window.
normalize : bool
If True, normalizes global closeness centrality for each year against the average closeness centrality for that year. This will require substantially more processing time, and values will usually be >> 0.
Returns : trajectory : dict
Global closeness centrality for node over specified period.