iranges: base structures and functions not unlike memoryviews¶
The module reflects the content of the R/Bioconductor package IRanges. It defines Python-level classes for the R/S4 classes, and gives otherwise access to R-level commands the usual rpy2:robjects way.
The variable iranges_env in the module is an rpy2.robjects.REnvironment for the modules namespace. Accessing explicitly a module’s object is then straightforward.
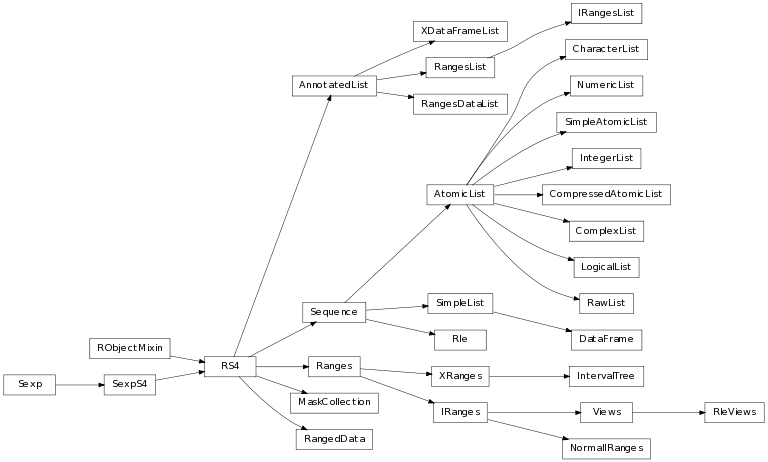
The class inheritance diagram presents the parent-child relationships between the elements.
A module to model the IRanges library in Bioconductor
Copyright 2009 - Laurent Gautier
- class bioc.iranges.AnnotatedList¶
- class bioc.iranges.AtomicList¶
- class bioc.iranges.CharacterList¶
- class bioc.iranges.ComplexList¶
- class bioc.iranges.CompressedAtomicList¶
- class bioc.iranges.DataFrame¶
- class bioc.iranges.IRanges¶
- class bioc.iranges.IRangesList¶
- class bioc.iranges.IntegerList¶
- class bioc.iranges.IntervalTree¶
- class bioc.iranges.LogicalList¶
- class bioc.iranges.NormalIRanges¶
- max¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- min¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- class bioc.iranges.NumericList¶
- class bioc.iranges.RangedData¶
- end¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- get_names()¶
- names¶
- start¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- width¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- class bioc.iranges.Ranges¶
- as_DataFrame()¶
- as_integer()¶
- as_matrix()¶
- end¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- isdisjoint()¶
- isempty()¶
- isnormal()¶
- mid()¶
- start¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- whichfirst_notnormal()¶
- width¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- class bioc.iranges.RangesDataList¶
- class bioc.iranges.RangesList¶
- class bioc.iranges.RawList¶
- class bioc.iranges.Rle¶
- class bioc.iranges.RleViews¶
- class bioc.iranges.Sequence¶
- Abstract class
- class bioc.iranges.SimpleAtomicList¶
- class bioc.iranges.SimpleList¶
- class bioc.iranges.XDataFrameList¶
- class bioc.iranges.XRanges¶
- bioc.iranges.iranges_conversion(robj)¶