geoquery: querying the GEO database¶
Example¶
import bioc.geoquery
gds = bioc.geoquery.__rpackage__.getGEO("GDS12")
>>> tuple(gds.meta.rx2("description"))
('Examination of two functionally distinct human bone marrow stromal cell lines, HS-27a and HS-5. Four independent RNA samples from each cell line analyzed.',
'HS-5',
'HS-27a')
Docstrings¶
The class inheritance diagram presents the parent-child relationships between the elements.
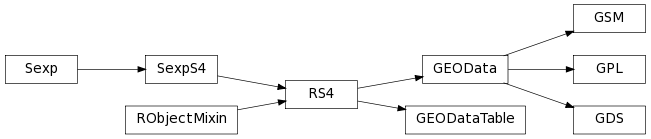
The module reflects some of the content of the R/Bioconductor package GEOquery. It defines Python-level classes for the R/S4 classes, and gives otherwise access to R-level commands the usual rpy2:robjects way.
In complement to this module, it is recommended to also use ‘rpy2.robjects.packages.importr’ to expose the full content of GEOquery
>>> import bioc.geoquery
Now all methods exposed by the R package are accessible from ‘bio.geoquery.__rpackage__’, with Python classes defined in bioc.geoquery mapping the R classes.
- class bioc.geoquery.GDS¶
- GEO ‘datasets’ entity
- class bioc.geoquery.GEOData¶
R ‘virtual’ class (that is an abstract class)
- accession¶
- GEO accession number
- columns¶
- column descriptions
- datatable¶
- data table
- meta¶
- meta-data (sample information)
- table¶
- table
- class bioc.geoquery.GPL¶
- GEO ‘platforms’ entity
- class bioc.geoquery.GSM¶
- GEO ‘samples’ entity
- bioc.geoquery.geoquery_conversion(robj)¶