gseabase: infrastructure for gene-set associations¶
The module reflects the content of the R/Bioconductor package GSEABase. It defines Python-level classes for the R/S4 classes, and gives otherwise access to R-level commands the usual rpy2:robjects way.
>>> import bioc.gseabase
XML description can be downloaded from the Broad Institute (http://www.broad.mit.edu/gsea/downloads.jsp)
>>> import from
>>> bioc.gseabase.__rpackage__.getBroadSets("")
The class inheritance diagram is useful for having an overview of how (biological) strings are modelled.
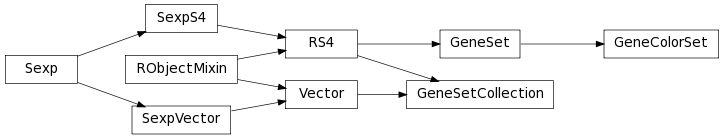
A module to model the GSEABase library in Bioconductor
Copyright 2009 - Laurent Gautier
- class bioc.gseabase.GeneColorSet¶
- coloring¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- genecolor¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- phenotype()¶
- phenotypecolor()¶
- rx(i)¶
- class bioc.gseabase.GeneSet¶
- collectiontype¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- contributor¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- creationdate¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- description¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- difference(gs)¶
- geneids¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- geneidtype¶
- maps GSEABase::geneIdType
- intersection(gs)¶
- longdescription¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- name¶
- maps GSEABase::setName
- organism¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- pubmedids¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- rx(i)¶
- union(gs)¶
- urls¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- class bioc.gseabase.GeneSetCollection(o)¶
- geneids¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- names¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- bioc.gseabase.gseabase_conversion(robj)¶