shortread: handle short reads¶
The module reflects the content of the R/Bioconductor package ShortRead. It defines Python-level classes for the R/S4 classes, and gives otherwise access to R-level commands the usual rpy2:robjects way.
Examples¶
>>> import bioc.shortread as shortread
>>> from rpy2.robjects.packages import importr
>>> base = importr('base')
Get the path to a FASTAQ file bundled with the R package. This is a small file, convenient to use as an example
>>> fp = base.system_file('extdata', package='ShortRead')
Now reading the data can used by calling directly the bioconductor functions.
>>> sr_pack = shortread.__rpackage__
>>> spath = sr_pack.SolexaPath(fp)
>>> rfq = sr_pack.readFastq(sr_pack.analysisPath(spath), pattern = "s_1_sequence.txt")
>>> rfq
<ShortReadQ - Python:0xca8376c / R:0xcd01cf8>
The class ShortReadQ has methods (mirroring the S4 methods in the R packages), and can be used as if an usual Python class.
For example, looking at the width for the first 10 reads:
>>> tuple(rfq.width[1:10])
(36, 36, 36, 36, 36, 36, 36, 36, 36)
Now on to reading aligned reads:
>>> aln = shortread.__rpackage__.readAligned(spath, "s_2_export.txt")
>>> aln
<AlignedRead - Python:0xce66a0c / R:0xd0c7f98>
Note
The function shortread.__rpackage__.readALined() can be specified the source of alignment data.
- bowtie: type = “Bowtie”
- MAQ: type=”MAQMapShort”, or type=”MAQMap”, or type=”MAQMapview”
Reads can be accessed:
>>> aln.do_slot('sread')
<DNAStringSet - Python:0xce7a0ac / R:0xd0c9484>
Note
In bioconductor, sread() is not formally declared as method of class AlignedRead, so we access it as a slot.
Filters can be used to extract subsets of the reads:
>>> # chromosome 5
>>> chromfilter = shortread.__rpackage__.chromosomeFilter('chr5.fa')
>>> aln_chr5 = aln.rx(chromfilter(aln))
>>> # position interval 6E6-10E6
>>> posfilter = shortread.__rpackage__.positionFilter(min=6E6, max=10E6)
>>> print(aln_chr5.rx(posfilter(aln_chr_5)))
class: AlignedRead
length: 1 reads; width: 35 cycles
chromosome: 1
position: 6774915
strand: 1
alignQuality: NumericQuality
alignData varLabels: run lane ... filtering contig
Docstrings¶
The class inheritance diagram presents the parent-child relationships between the elements.
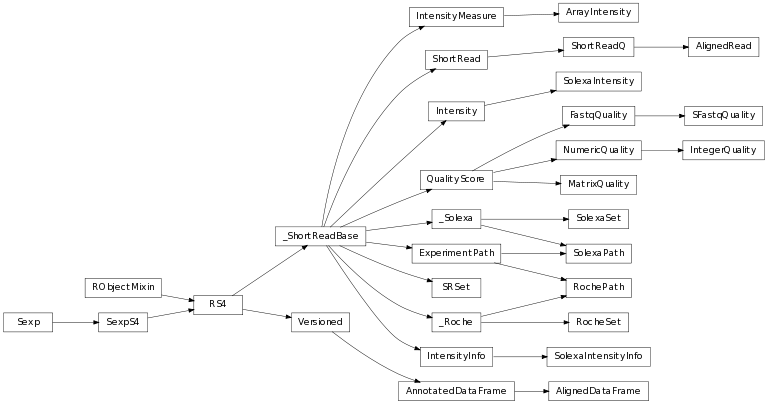
A module to model the ShortRead library in Bioconductor
Copyright 2009 - Laurent Gautier
- class bioc.shortread.AlignedRead¶
- coverage(start=NA, end=NA, coords=<StrVector - Python:0x59926e8 / R:0x909df08>, extend=0)¶
- rx(i, drop=True)¶
- class bioc.shortread.ArrayIntensity¶
- class bioc.shortread.FastqQuality¶
- alphabetbycycle()¶
- alphabetfrequency()¶
- srduplicated()¶
- srorder()¶
- srrank()¶
- class bioc.shortread.IntegerQuality¶
- class bioc.shortread.Intensity¶
- class bioc.shortread.IntensityInfo¶
- class bioc.shortread.IntensityMeasure¶
- class bioc.shortread.MatrixQuality¶
- class bioc.shortread.NumericQuality¶
- class bioc.shortread.QualityScore¶
- detail()¶
- width¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- class bioc.shortread.RochePath¶
- detail()¶
- read454()¶
- readfasta(**kwargs)¶
- readpath()¶
- readqual(**kwargs)¶
- rocheset()¶
- runnames()¶
- class bioc.shortread.RocheSet¶
- class bioc.shortread.SFastqQuality¶
- class bioc.shortread.SRSet¶
- class bioc.shortread.ShortRead¶
- alphabetbycycle()¶
- clean()¶
- detail()¶
- narrow(start=None, end=None, width=None, use_names=True)¶
- srduplicated()¶
- srorder()¶
- srrank()¶
- srsort()¶
- tables(n=50)¶
- width¶
- Python representation of an R function such as the character ‘.’ is replaced with ‘_’ whenever present in the R argument name.
- class bioc.shortread.ShortReadQ¶
- class bioc.shortread.SolexaIntensity¶
- class bioc.shortread.SolexaIntensityInfo¶
- class bioc.shortread.SolexaPath¶
- detail()¶
- readaligned(**kwargs)¶
- readfastq(**kwargs)¶
- readintensities(**kwargs)¶
- readprb(**kwargs)¶
- readqseq(**kwargs)¶
- solexaset()¶
- class bioc.shortread.SolexaSet¶
- bioc.shortread.shortread_conversion(robj)¶