Rosetta¶
Contents:
Indices and tables¶
All Modules and Classes¶
cmd¶
Unix-like command line utilities. Filters (read from stdin/write to stdout) for files
Installation should put these in your path. To see help, do
module_name.py -h
cut¶
subsample¶
split¶
row_filter¶
files_to_vw¶
join_csv¶
concat_csv¶
parallel¶
- Wrappers for Python multiprocessing that add ease of use
- Memory-friendly multiprocessing
parallel_easy¶
Functions to assist in parallel processing with Python 2.7.
- Memory-friendly iterator functionality (wrapping Pool.imap).
- Exit with Ctrl-C.
- Easy use of n_jobs. (e.g. when n_jobs == 1, processing is serial)
- Similar to joblib.Parallel but with the addition of imap functionality and a more effective way of handling Ctrl-C exit (we add a timeout).
-
rosetta.parallel.parallel_easy.imap_easy(func, iterable, n_jobs, chunksize, ordered=True)[source]¶ Returns a parallel iterator of func over iterable.
Worker processes return one “chunk” of data at a time, and the iterator allows you to deal with each chunk as they come back, so memory can be handled efficiently.
Parameters: func : Function of one variable
You can use functools.partial to build this. A lambda function will not work
iterable : List, iterator, etc...
func is applied to this
n_jobs : Integer
The number of jobs to use for the computation. If -1 all CPUs are used. If 1 is given, no parallel computing code is used at all, which is useful for debugging. For n_jobs below -1, (n_cpus + 1 + n_jobs) are used. Thus for n_jobs = -2, all CPUs but one are used.
chunksize : Integer
Jobs/results will be sent between master/slave processes in chunks of size chunksize. If chunksize is too small, communication overhead slows things down. If chunksize is too large, one process ends up doing too much work (and large results will up in memory).
ordered : Boolean
If True, results are dished out in the order corresponding to iterable. If False, results are dished out in whatever order workers return them.
Examples
>>> from functools import partial >>> from rosetta.parallel.parallel_easy import imap_easy >>> def abfunc(x, a, b=1): ... return x * a * b >>> some_numbers = range(3) >>> func = partial(abfunc, 2, b=3) >>> results_iterator = imap_easy(func, some_numbers, 2, 5) >>> for result in results_iterator: ... print result 0 6 12
-
rosetta.parallel.parallel_easy.map_easy(func, iterable, n_jobs)[source]¶ Returns a parallel map of func over iterable. Returns all results at once, so if results are big memory issues may arise
Parameters: func : Function of one variable
You can use functools.partial to build this. A lambda function will not work
iterable : List, iterator, etc...
func is applied to this
n_jobs : Integer
The number of jobs to use for the computation. If -1 all CPUs are used. If 1 is given, no parallel computing code is used at all, which is useful for debugging. For n_jobs below -1, (n_cpus + 1 + n_jobs) are used. Thus for n_jobs = -2, all CPUs but one are used.
Examples
>>> from functools import partial >>> from rosetta.parallel.parallel_easy import map_easy >>> def abfunc(x, a, b=1): ... return x * a * b >>> some_numbers = range(5) >>> func = partial(abfunc, 2, b=3) >>> map_easy(func, some_numbers) [0, 6, 12, 18, 24]
-
rosetta.parallel.parallel_easy.map_easy_padded_blocks(func, iterable, n_jobs, pad, blocksize=None)[source]¶ Returns a parallel map of func over iterable, computed by splitting iterable into padded blocks, then piecing the result together.
Parameters: func : Function of one variable
You can use functools.partial to build this. A lambda function will not work
iterable : List, iterator, etc...
func is applied to this
n_jobs : Integer
The number of jobs to use for the computation. If -1 all CPUs are used. If 1 is given, no parallel computing code is used at all, which is useful for debugging. For n_jobs below -1, (n_cpus + 1 + n_jobs) are used. Thus for n_jobs = -2, all CPUs but one are used.
pad : Nonnegative Integer
Each block is processed with pad extra on each side.
blocksize : Nonnegative Integer
If None, use 100 * pad
Returns: result : List
Equivalent to list(func(iterable))
Examples
>>> numbers = [0, 0, 2, -1, 4, 2, 6, 7, 6, 9] >>> pad = 1 >>> n_jobs = -1 >>> def rightmax(mylist): ... return [max(mylist[i: i+2]) for i in range(len(mylist))] >>> result = map_easy_padded_blocks(rightmax, numbers, n_jobs, pad) >>> benchmark = rightmax(numbers) >>> result == benchmark True
pandas_easy¶
Functions for helping make pandas parallel.
-
rosetta.parallel.pandas_easy.groupby_to_scalar_to_series(df_or_series, func, n_jobs, **groupby_kwargs)[source]¶ Returns a parallelized, simplified, and restricted version of: df_or_series.groupby(**groupby_kwargs).apply(func)
Works ONLY for the simple case that .apply(func) would yield a Series of length equal to the number of groups, in other words, func applied to each group is a scalar.
Parameters: df_or_series : DataFrame or Series
This is what is grouped
func : Function
Applied to each group using func(df_or_series) Should return one single value (e.g. string or number) Must be picklable: A lambda function will not work!
groupby_kwargs : Keyword args
Passed directly to DataFrame.groupby to determine groups. The most common one is “by”, e.g.
by=’a’ by=my_grouper_function by=my_grouping_list_of_labels
Returns: result : Series
Index is the group names Values are func(group) iterated over every group
Examples
>>> from rosetta.parallel.pandas_easy import groupby_to_series >>> df = pd.DataFrame({'a': [6, 2, 2], 'b': [4, 5, 6]}) >>> df a b 0 6 4 1 2 5 2 2 6 >>> groupby_to_series(df, max, n_jobs, by='a') 2 b 6 b
>>> s = pd.Series([1, 2, 3, 4]) >>> s 0 1 1 2 2 3 3 4 >>> labels = ['a', 'a', 'b', 'b'] >>> groupby_to_series(s, max, 1, by=labels) a 2 b 4
-
rosetta.parallel.pandas_easy.groupby_to_series_to_frame(frame, func, n_jobs, use_apply=True, **groupby_kwargs)[source]¶ A parallel function somewhat similar DataFrame.groupby.apply(func).
For each group in df_or_series.groupby(**groupby_kwargs), compute func(group) or group.apply(func) and, assuming each result is a series, flatten each series then paste them together.
Parameters: frame : DataFrame
func : Function
Applied to each group using func(df_or_series) Must be picklable: A lambda function will not work!
use_apply : Boolean
If True, use group.apply(func) If False, use func(group)
groupby_kwargs : Keyword args
Passed directly to DataFrame.groupby to determine groups. The most common one is “by”, e.g.
by=’a’ by=my_grouper_function by=my_grouping_list_of_labels
Returns: result : DataFrame
Index is the group names Values are func(group) iterated over every group, then pasted together
Examples
>>> from rosetta.parallel.pandas_easy import groupby_to_series_to_frame >>> df = pd.DataFrame({'a': [6, 2, 2], 'b': [4, 5, 6]}) >>> labels = ['g1', 'g1', 'g2'] # Result and benchmark will be equal...despite the fact that you can't # do df.groupby(labels).apply(np.mean) >>> benchmark = df.groupby(labels).mean() >>> result = groupby_to_series_to_frame( ... df, np.mean, 1, use_apply=True, by=labels) >>> print result a b g1 4 4.5 g2 2 6.0
text¶
Text-processing specific
- Stream text from disk to formats used in common ML processes
- Write processed text to sparse formats
- Helpers for ML tools (e.g. Vowpal Wabbit, Gensim, etc...)
- Other general utilities
filefilter¶
Contains a collection of function that clean, decode and move files around.
-
class
rosetta.text.filefilter.PathFinder(text_base_path=None, file_type='*', name_strip='\..*', limit=None)[source]¶ Find and access paths in a directory tree.
-
rosetta.text.filefilter.get_paths(base_path, file_type='*', relative=False, get_iter=False, limit=None)[source]¶ Crawls subdirectories and returns an iterator over paths to files that match the file_type.
Parameters: base_path : String
Path to the directory that will be crawled
file_type : String
String to filter files with. E.g. ‘*.txt’. Note that the filenames will be converted to lowercase before this comparison.
relative : Boolean
If True, get paths relative to base_path If False, get absolute paths
get_iter : Boolean
If True, return an iterator over paths rather than a list.
-
rosetta.text.filefilter.path_to_name(path, strip_ext=True)[source]¶ Takes one path and returns the filename, excluding the extension.
-
rosetta.text.filefilter.path_to_newname(path, name_level=1)[source]¶ Takes one path and returns a new name, combining the directory structure with the filename.
Parameters: path : String
name_level : Integer
Form the name using items this far back in the path. E.g. if path = mydata/1234/3.txt and name_level == 2, then name = 1234_3
Returns: name : String
streamers¶
Classes for streaming tokens/info from files/sparse files etc...
-
class
rosetta.text.streamers.BaseStreamer[source]¶ Base class...don’t use this directly.
Methods
-
info_stream(**kwargs)[source]¶ Abstract method. All derived classes will implement to return an interator over the text documents with processing as appropriate.
-
single_stream(item, cache_list=None, **kwargs)[source]¶ Stream a single item from source.
Parameters: item : String
The single item to pull from info and stream.
cache_list : List of strings
Cache these items on every iteration
kwargs : Keyword args
Passed on to self.info_stream
-
to_scipysparse(cache_list=None, **kwargs)[source]¶ Returns a scipy sparse matrix representing the collection of documents as a bag of words, one (sparse) row per document. Translation from column to token are stored into the cache
Parameters: cache_list : List of strings.
Cache these items as they appear E.g. self.token_stream(‘doc_id’, ‘tokens’) caches info[‘doc_id’] and info[‘tokens’] (assuming both are available).
kwargs : Keyword args
Passed on to self.info_stream
-
to_vw(out_stream=<open file '<stdout>', mode 'w'>, n_jobs=1, raise_on_bad_id=True, cache_list=None)[source]¶ Write our filestream to a VW (Vowpal Wabbit) formatted file.
Parameters: out_stream : stream, buffer or open file object
n_jobs : Integer
number of CPU jobs
cache_list : List of strings.
Cache these items as they appear E.g. self.token_stream(‘doc_id’, ‘tokens’) caches info[‘doc_id’] and info[‘tokens’] (assuming both are available).
Notes: :
—– :
self.info_stream must have a ‘doc_id’ field, as this is used to index :
the lines in the vw file. :
-
token_stream(cache_list=None, **kwargs)[source]¶ Returns an iterator over tokens with possible caching of other info.
Parameters: cache_list : List of strings.
Cache these items as they appear E.g. self.token_stream(‘doc_id’, ‘tokens’) caches info[‘doc_id’] and info[‘tokens’] (assuming both are available).
kwargs : Keyword args
Passed on to self.info_stream
-
-
class
rosetta.text.streamers.DBStreamer(db_setup, tokenizer=None, tokenizer_func=None)[source]¶ Database streamer base class
Methods
-
iterate_over_query()[source]¶ Return an iterator over query result. We suggest that the entire query result not be returned and that iteration is controlled on server side, but this method does not guarantee that. This method must return a dictionary, which at least has the key ‘text’ in it, containing the next to be tokenized.
-
-
class
rosetta.text.streamers.MongoStreamer(db_setup, tokenizer=None, tokenizer_func=None)[source]¶ Subclass of DBStreamer to connect to a Mongo database and iterate over query results. db_setup is expected to be a dictionary containing host, database, collection, query, and text_key. Additionally an optional limit parameter is allowed. The query itself must return a column named text_key which is passed on as ‘text’ to the iterator. In addition, because it is difficult to rename mongo fields (similar to the SQL ‘AS’ syntax), we allow a translation dictionary to be passed in, which translates keys in the mongo dictionary result names k to be passed into the result as v for key value pairs {k : v}. Currently we don’t deal with nested documents.
Example:
db_setup = {} db_setup[‘host’] = ‘localhost’ db_setup[‘database’] = ‘places’ db_setup[‘collection’] = ‘opentable’ db_setup[‘query’] = {} db_setup[‘limit’] = 5 db_setup[‘text_key’] = ‘desc’ db_setup[‘translations’] = {‘_id’ : ‘doc_id’}
# In this example, we assume that the collection has a field named # desc, holding the text to be analyzed, and a field named _id which # will be translated to doc_id and stored in the cache.
my_tokenizer = TokenizerBasic() stream = MongoStreamer(db_setup=db_setup, tokenizer=my_tokenizer)
- for text in stream.info_stream(cache_list=[‘doc_id’]):
- print text[‘doc_id’], text[‘tokens’]
Methods
-
class
rosetta.text.streamers.MySQLStreamer(*args, **kwargs)[source]¶ Subclass of DBStreamer to connect to a MySQL database and iterate over query results. db_setup is expected to be a dictionary containing host, user, password, database, and query. The query itself must return a column named text.
- Example:
db_setup = {} db_setup[‘host’] = ‘hostname’ db_setup[‘user’] = ‘username’ db_setup[‘password’] = ‘password’ db_setup[‘database’] = ‘database’ db_setup[‘query’] = ‘select
id as doc_id, body as textfrom tablename where length(body) > 100’
my_tokenizer = TokenizerBasic() stream = MySQLStreamer(db_setup=db_setup, tokenizer=my_tokenizer)
- for text in stream.info_stream(cache_list=[‘doc_id’]):
- print text[‘doc_id’], text[‘tokens’]
Methods
-
class
rosetta.text.streamers.TextFileStreamer(text_base_path=None, path_list=None, file_type='*', name_strip='\..*', tokenizer=None, tokenizer_func=None, limit=None, shuffle=True)[source]¶ For streaming from text files.
Methods
-
info_stream(paths=None, doc_id=None, limit=None)[source]¶ Returns an iterator over paths yielding dictionaries with information about the file contained within.
Parameters: paths : list of strings
doc_id : list of strings or ints
limit : Integer
Use limit in place of self.limit.
-
to_vw(outfile, n_jobs=1, chunksize=1000, raise_on_bad_id=True)[source]¶ Write our filestream to a VW (Vowpal Wabbit) formatted file.
Parameters: outfile : filepath or buffer
n_jobs : Integer
Use n_jobs different jobs to do the processing. Set = 4 for 4 jobs. Set = -1 to use all available, -2 for all except 1,...
chunksize : Integer
Workers process this many jobs at once before pickling and sending results to master. If this is too low, communication overhead will dominate. If this is too high, jobs will not be distributed evenly.
raise_on_bad_id : Boolean
If True, raise DocIDError when the doc_id (formed by self) is not a valid VW “Tag”. I.e. contains :, |, ‘, or whitespace. If False, print warning.
-
-
class
rosetta.text.streamers.TextIterStreamer(text_iter, tokenizer=None, tokenizer_func=None)[source]¶ For streaming text.
Methods
-
class
rosetta.text.streamers.VWStreamer(sfile=None, cache_sfile=False, limit=None, shuffle=False)[source]¶ For streaming from a single VW file. Since the VW file format does not preserve token order, all tokens are unordered.
Methods
text_processors¶
Tokenizer¶
Classes with a .text_to_token_list method (and a bit more). Used by other modules as a means to convert stings to lists of strings.
If you have a function that converts strings to lists of strings, you can make a tokenizer from it by using MakeTokenizer(my_tokenizing_func).
SparseFormatter¶
Classes for converting text to sparse representations (e.g. VW or SVMLight).
SFileFilter¶
Classes for filtering words/rows from a sparse formatted file.
-
class
rosetta.text.text_processors.MakeTokenizer(tokenizer_func)[source]¶ Makes a subclass of BaseTokenizer out of a function.
Methods
-
class
rosetta.text.text_processors.SFileFilter(formatter, bit_precision=18, sfile=None, verbose=True)[source]¶ Filters results stored in sfiles (sparsely formattted bag-of-words files).
Methods
-
compactify()[source]¶ Removes “gaps” in the id values in self.token2id. Every single id value will (probably) be altered.
-
filter_extremes(doc_freq_min=0, doc_freq_max=inf, doc_fraction_min=0, doc_fraction_max=1, token_score_min=0, token_score_max=inf, token_score_quantile_min=0, token_score_quantile_max=1)[source]¶ Remove extreme tokens from self (calling self.filter_tokens).
Parameters: doc_freq_min : Integer
Remove tokens that in less than this number of documents
doc_freq_max : Integer
doc_fraction_min : Float in [0, 1]
Remove tokens that are in less than this fraction of documents
doc_fraction_max : Float in [0, 1]
token_score_quantile_min : Float in [0, 1]
Minimum quantile that the token score (usually total token count) can be in.
token_score_quantile_max : Float in [0, 1]
Maximum quantile that the token score can be in
Returns: self :
-
filter_sfile(infile, outfile, doc_id_list=None, enforce_all_doc_id=True, min_tf_idf=0, filters=None)[source]¶ Alter an sfile by converting tokens to id values, and removing tokens not in self.token2id. Optionally filters on doc_id, tf_idf and user-defined filters.
Parameters: infile : file path or buffer
outfile : file path or buffer
doc_id_list : Iterable over strings
Keep only rows with doc_id in this list
enforce_all_doc_id : Boolean
If True (and doc_id is not None), raise exception unless all doc_id in doc_id_list are seen.
min_tf_idf : int or float
Keep only tokens whose term frequency-inverse document frequency is greater than this threshold. Given a token t and a document d in a corpus of documents D, tf_idf is given by the following formula:
tf_idf(t, d, D) = tf(t, d) x idf(t, D),
- where
- tf(t, d) is the number of times the term t shows up in the document d,
- idf(t, D) = log (N / M), where N is the total number of documents in D and M is the number of documents in D which contain the token t. The logarithm is base e.
filters : iterable over functions
Each function must take a record_dict as a parameter and return a boolean. The record_dict may (and usually should) be altered in place. If the return value is False, the record_dict (corresponding to a document) is filtered out of the sfile. Both the doc_id_list and min_tf_idf parameters are implemented in this style internally. If the doc_id_list or min_tf_idf flags are set, those filters will run before the those found in filters. See
rosetta/text/streaming_filters.py
in the rosetta repository for the implementation details of the record_dict and built-in filters as well as explanations of how to define more filters.
-
filter_tokens(tokens)[source]¶ Remove tokens from appropriate attributes.
Parameters: tokens : String or iterable over strings
E.g. a single token or list of tokens
Returns: self :
-
load_sfile(sfile)[source]¶ Load an sfile, building self.token2id
Parameters: sfile : String or open file
The sparse formatted file we will load.
Returns: self :
-
save(savepath, protocol=-1, set_id2token=True)[source]¶ Pickle self to outfile.
Parameters: savefile : filepath or buffer
protocol : 0, 1, 2, -1
0 < 1 < 2 in terms of performance. -1 means use highest available.
set_id2token : Boolean
If True, set self.id2token before saving. Used to associate tokens with the output of a VW file.
-
set_bit_precision_required()[source]¶ Sets self.bit_precision_required to the minimum bit precision b such that all token id values are less than 2^b.
The idea is that only compactification can change this, so we only (automatically) call this after compactification.
-
-
class
rosetta.text.text_processors.SVMLightFormatter[source]¶ For formatting in/out of SVM-Light format (info not currently supported) http://svmlight.joachims.org/
<line> .=. <target> <feature>:<value> <feature>:<value> ... <target> .=. +1 | -1 | 0 | <float> <feature> .=. <integer> | “qid” <value> .=. <float> <info> .=. <string>
Methods
-
get_sstr(feature_values=None, target=1, importance=None, doc_id=None)[source]¶ Return a string reprsenting one record in SVM-Light sparse format <line> .=. <target> <feature>:<value> <feature>:<value>
Parameters: feature_values : Dict-like
{hash1: value1,...}
target : Real number
The value we are trying to predict.
Returns: formatted : String
Formatted in SVM-Light
-
-
class
rosetta.text.text_processors.SparseFormatter[source]¶ Base class for sparse formatting, e.g. VW or svmlight. Not meant to be directly used.
Methods
-
sfile_to_token_iter(filepath_or_buffer, limit=None)[source]¶ Return an iterator over filepath_or_buffer that returns, line-by-line, a token_list.
Parameters: filepath_or_buffer : string or file handle / StringIO.
File should be formatted according to self.format.
Returns: token_iter : Iterator
E.g. token_iter.next() gets the next line as a list of tokens.
-
sstr_to_dict(sstr)[source]¶ Returns a dict representation of sparse record string.
Parameters: sstr : String
String representation of one record.
Returns: record_dict : Dict
possible keys = ‘target’, ‘importance’, ‘doc_id’, ‘feature_values’
Notes
rstrips newline characters from sstr before parsing.
-
sstr_to_info(sstr)[source]¶ Returns the full info dictionary corresponding to a sparse record string. This holds “everything.”
Parameters: sstr : String
String representation of one record.
Returns: info : Dict
- possible keys = ‘tokens’, ‘target’, ‘importance’, ‘doc_id’,
‘feature_values’, etc...
-
sstr_to_token_list(sstr)[source]¶ Convertes a sparse record string to a list of tokens (with repeats) corresponding to sstr.
E.g. if sstr represented the dict {‘hi’: 2, ‘bye’: 1}, then token_list = [‘hi’, ‘hi’, ‘bye’] (up to permutation).
Parameters: sstr : String
Formatted according to self.format_name Note that the values in sstr must be integers.
Returns: token_list : List of Strings
-
-
class
rosetta.text.text_processors.TokenizerBasic[source]¶ A simple tokenizer. Extracts word counts from text.
Keeps only non-stopwords, converts to lowercase, keeps words of length >=2.
Methods
-
class
rosetta.text.text_processors.TokenizerPOSFilter(pos_types=[], sent_tokenizer=<function sent_tokenize>, word_tokenizer=<rosetta.text.text_processors.TokenizerBasic object>, word_tokenizer_func=None, pos_tagger=<function pos_tag>)[source]¶ Tokenizes, does POS tagging, then keeps words that match particular POS.
Methods
-
class
rosetta.text.text_processors.VWFormatter[source]¶ Converts in and out of VW format (namespaces currently not supported). Many valid VW inputs are possible, we ONLY support
[target] [Importance [Tag]]| feature1[:value1] feature2[:value2] ...
Every single whitespace, pipe, colon, and newline is significant.
See: https://github.com/JohnLangford/vowpal_wabbit/wiki/Input-format http://hunch.net/~vw/validate.html
Methods
-
get_sstr(feature_values=None, target=None, importance=None, doc_id=None)[source]¶ Return a string reprsenting one record in sparse VW format:
Parameters: feature_values : Dict-like
{feature1: value1,...}
target : Real number
The value we are trying to predict.
importance : Real number
The importance weight to associate to this example.
doc_id : Number or string
A name for this example.
Returns: formatted : String
Formatted in VW format
-
-
rosetta.text.text_processors.collision_probability(vocab_size, bit_precision)[source]¶ Approximate probability of at least one collision (assuming perfect hashing). See the Wikipedia article on “The birthday problem” for details.
Parameters: vocab_size : Integer
Number of unique words in vocabulary
bit_precision : Integer
Number of bits in space we are hashing to
nlp¶
-
rosetta.text.nlp.bigram_tokenize(text, word_tok=<function word_tokenize>, skip_regex='\\.|, |:|;|\\?|!', **word_tok_kwargs)[source]¶ Same as bigram_tokenize_iter, except returns a list.
Bigram tokenizer generator function.
vw_helpers¶
Wrappers to help with Vowpal Wabbit (VW).
-
class
rosetta.text.vw_helpers.LDAResults(topics_file, predictions_file, sfile_filter, num_topics=None, alpha=None, verbose=False)[source]¶ Facilitates working with results of VW lda runs. Only useful when you’re following the workflow outlined here:
https://github.com/columbia-applied-data-science/rosetta/blob/master/examples/vw_helpers.md
Methods
-
cosine_similarity(frame1, frame2)[source]¶ Computes doc-doc similarity between rows of two frames containing document topic weights.
Parameters: frame1, frame2 : DataFrame or Series
Rows are different records, columns are topic weights. self.pr_topic_g_doc is an example of a (large) frame of this type.
Returns: sims : DataFrame
sims.ix[i, j] is similarity between frame1[i] and frame2[j]
-
predict(tokenized_text, maxiter=50, atol=0.001, raise_on_unknown=False)[source]¶ Returns a probability distribution over topics given that one (tokenized) document is equal to tokenized_text.
This is NOT equivalent to prob_token_topic(c_token=tokenized_text), since that is an OR statement about the tokens, and this is an AND.
Parameters: tokenized_text : List of strings
Represents the tokens that are in some document text.
maxiter : Integer
Maximum iterations used in updating parameters.
atol : Float
Absolute tolerance for change in parameters before converged.
raise_on_unknown : Boolean
If True, raise TokenError when all tokens are unknown to this model.
Returns: prob_topics : Series
self.pr_topic_g_doc is an example of a (large) frame of this type.
Notes
Treats this as a new document and figures out topic weights for it using the existing token-topic weights. Does NOT update previous results/weights.
-
print_topics(num_words=5, outfile=<open file '<stdout>', mode 'w'>, show_doc_fraction=True)[source]¶ Print the top results for self.pr_token_g_topic for all topics
Parameters: num_words : Integer
Print the num_words words (ordered by P[w|topic]) in each topic.
outfile : filepath or buffer
Write results to this file.
show_doc_fraction : Boolean
If True, print doc_fraction along with the topic weight
-
prob_doc_topic(doc=None, topic=None, c_doc=None, c_topic=None)[source]¶ Return joint probabilities of (doc, topic), restricted to subsets, conditioned on variables.
Parameters: doc : list-like or string
Restrict returned probabilities to these doc_ids
topic : list-like or string
Restrict returned probabilities to these topics
c_doc : list-like or string
Condition on doc_id in c_doc
c_topic : list-like or string
Condition on topic in c_topic
Examples
- prob_doc_topic(c_topic=[‘topic_0’])
- = P(doc, topic | topic in [‘topic_0’]) for all possible (doc, topic) pairs
- prob_doc_topic(doc=[‘doc0’, ‘doc1’], c_topic=[‘topic_0’])
- = P(doc, topic | topic in [‘topic_0’]) for all (doc, topic) pairs with doc in [‘doc0’, ‘doc1’]
- prob_doc_topic(doc=[‘doc0’, ‘doc1’], topic=[‘topic_0’])
- = P(doc, topic) for all (doc, topic) pairs with doc in [‘doc0’, ‘doc1’] and topic in [‘topic_0’]
-
prob_token_topic(token=None, topic=None, c_token=None, c_topic=None)[source]¶ Return joint densities of (token, topic), restricted to subsets, conditioned on variables.
Parameters: token : list-like or string
Restrict returned probabilities to these tokens
topic : list-like or string
Restrict returned probabilities to these topics
c_token : list-like or string
Condition on token in c_token
c_topic : list-like or string
Condition on topic in c_topic
Examples
- prob_token_topic(c_topic=[‘topic_0’])
- = P(token, topic | topic in [‘topic_0’]) for all possible (token, topic) pairs
- prob_token_topic(token=[‘war’, ‘peace’], c_topic=[‘topic_0’])
- = P(token, topic | topic in [‘topic_0’]) for all (token, topic) pairs with token in [‘war’, ‘peace]
- prob_token_topic(token=[‘war’, ‘peace’], topic=[‘topic_0’])
- = P(token, topic) for all (token, topic) pairs with token in [‘war’, ‘peace] and topic in [‘topic_0’]
-
-
rosetta.text.vw_helpers.find_start_line_lda_predictions(predictions_file, num_topics)[source]¶ Return the line number (zero indexed) of the start of the last set of predictions in predictions_file.
Parameters: predictions_file : filepath or buffer
The -p output of a VW lda run
num_topics : Integer
The number of topics you should see
Notes
The predictions_file contains repeated predictions...one for every pass. We parse out and include only the last predictions by looking for repeats of the first lines doc_id field. We thus, at this time, require the VW formatted file to have, in the last column, a unique doc_id associated with the doc.
-
rosetta.text.vw_helpers.parse_lda_predictions(predictions_file, num_topics, start_line, normalize=True, get_iter=False)[source]¶ Return a DataFrame representation of a VW prediction file.
Parameters: predictions_file : filepath or buffer
The -p output of a VW lda run
num_topics : Integer
The number of topics you should see
start_line : Integer
Start reading the predictions file here. The predictions file contains repeated predictions, one for every pass. You generally do not want every prediction.
normalize : Boolean
Normalize the rows so that they represent probabilities of topic given doc_id.
get_iter : Boolean
if True will return a iterator yielding dict of doc_id and topic probs
-
rosetta.text.vw_helpers.parse_lda_topics(topics_file, num_topics, max_token_hash=None, normalize=True, get_iter=False)[source]¶ Returns a DataFrame representation of the topics output of an lda VW run.
Parameters: topics_file : filepath or buffer
The –readable_model output of a VW lda run
num_topics : Integer
The number of topics in every valid row
max_token_hash : Integer
Reading of token probabilities from the topics_file will ignore all token with hash above this value. Useful, when you know the max hash value of your tokens.
normalize : Boolean
Normalize the rows of the data frame so that they represent probabilities of topic given hash_val.
get_iter : Boolean
if True will return a iterator yielding dict of hash and token vals
Notes
The trick is dealing with lack of a marker for the information printed on top, and the inconsistant delimiter choice.
gensim_helpers¶
Helper objects/functions specifically for use with Gensim.
-
class
rosetta.text.gensim_helpers.StreamerCorpus(streamer, dictionary, doc_id=None, limit=None)[source]¶ A “corpus type” object built with token streams and dictionaries.
Depending on your method for streaming tokens, this could be slow... Before modeling, it’s usually better to serialize this corpus using:
self.to_corpus_plus(fname) or gensim.corpora.SvmLightCorpus.serialize(path, self)
Methods
-
class
rosetta.text.gensim_helpers.SvmLightPlusCorpus(fname, doc_id=None, doc_id_filter=None, limit=None)[source]¶ Extends gensim.corpora.SvmLightCorpus, providing methods to work with (e.g. filter by) doc_ids.
Methods
-
classmethod
from_streamer_dict(streamer, dictionary, fname, doc_id=None, limit=None)[source]¶ Initialize from a Streamer and gensim.corpora.dictionary, serializing the corpus (to disk) in SvmLightPlus format, then returning a SvmLightPlusCorpus.
Parameters: streamer : Streamer compatible object.
Method streamer.token_stream() returns a stream of lists of words.
dictionary : gensim.corpora.Dictionary object
fname : String
Path to save the bag-of-words file at
doc_id : Iterable over strings
Limit all streaming results to docs with these doc_ids
limit : Integer
Limit all streaming results to this many
Returns: corpus : SvmLightCorpus
-
classmethod
modeling¶
- General ML modeling utilities
eda¶
-
rosetta.modeling.eda.get_labels(series, bins=10, quantiles=False)[source]¶ Divides series into bins and returns labels corresponding to midpoints of bins.
Parameters: series : Pandas.Series of numeric data
bins : Positive Integer, optional
Number of bins to divide series
quantiles : Boolean, optional
If True, bin data using quantiles rather than an evenly divided range
-
rosetta.modeling.eda.hist_cols(df, cols_to_plot, num_cols, num_rows, figsize=None, **kwargs)[source]¶ Plots histograms of columns of a DataFrame as subplots in one big plot. Handles nans and extreme values in a “graceful” manner by removing them and reporting their occurance.
Parameters: df : Pandas DataFrame
cols_to_plot : List
Column names of df that will be plotted
num_cols, num_rows : Positive integers
Number of columns and rows in the plot
figsize : (x, y) tuple, optional
Size of the figure
**kwargs : Keyword args to pass on to plot
-
rosetta.modeling.eda.hist_one_col(col)[source]¶ Plots a histogram one column. Handles nans and extreme values in a “graceful” manner.
-
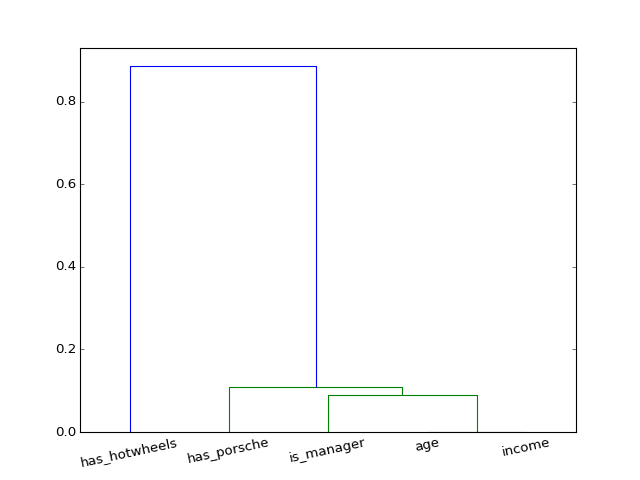
rosetta.modeling.eda.plot_corr_dendrogram(corr, cluster_method='weighted', **dendrogram_kwargs)[source]¶ Plot a correlation matrix as a dendrogram (on the current axes). Uses scipy.cluster.hierarchy.linkage to compute clusters based on distance between samples.
Since correlation is passed in, this correlation must be converted to a distance (using distance_fun). The default distance_fun makes highly correlated points have low distance, and vice versa.
Parameters: corr : numpy ndarray or pandas DataFrame
corr[i, j] is the correlation (should be between -1 and 1) of samples i and j.
cluster_method : String
Method to use to amalgomate clusters. Either ‘single’, ‘complete’, ‘average’, or ‘weighted’. See scipy.cluster.hierarchy.linkage for details.
dendrogram_kwargs : Additional kwargs
Pass to the call of scipy.cluster.hierarchy.dendrogram()
-
rosetta.modeling.eda.plot_corr_grid(corr, cluster=True, cluster_method='weighted', distance_fun=None, ax=None, **fig_kwargs)[source]¶ Plot a correlation matrix as a grid. Uses scipy.cluster.hierarchy.linkage to compute clusters based on distance between samples.
Since correlation is passed in, this correlation must be converted to a distance (using distance_fun). The default distance_fun makes highly correlated points have low distance, and vice versa.
Parameters: corr : numpy ndarray or pandas DataFrame
corr[i, j] is the correlation (should be between -1 and 1) of samples i and j.
cluster : Boolean
If True, reorder the matrix putting correlated entries nearby.
distance_fun : Function
inter-variable distance = distance_fun(corr). If None, use (1 - corr) / 2.
ax : matplotlib AxesSubplot instance
If None, use pl.gca()
cluster_method : String
Method to use to amalgomate clusters. Either ‘single’, ‘complete’, ‘average’, or ‘weighted’. See scipy.cluster.hierarchy.linkage for details.
Returns: fig : matplotlib figure instance
-
rosetta.modeling.eda.plot_reducedY_vs_binnedX(x, y, Y_reducer=<function mean>, X_reducer=<function mean>, bins=10, quantiles=False, plot_count_X=False, **plt_kwargs)[source]¶ Bin X and, inside every bin, apply Y_reducer to the Y values. Then plot.
Parameters: x : Pandas.Series with numeric data
y : Pandas.Series with numeric data = 0 or 1
Y_reducer : function
Used to aggregate the Y values in every bin
X_reducer : function
Used to aggregate the X values in every bin. This gives us the bin labels that are used as the indices.
bins : Positive Integer, optional
Number of bins to divide series
quantiles : Boolean, optional
If True, bin data using quantiles rather than an evenly divided range
plot_count_X : Boolean, optional
If True, plot count_X versus x in a separate subplot
**kwargs : Extra keywordargs passed to plot
Examples
Suppose Y is binary. Then to plot P[Y=1|X=x] (for x inside the bins), as well as #[X=x], use:
eda.plot_reducedY_vs_binnedX(x, y, Y_reducer=np.mean, plot_count_X=True)
-
rosetta.modeling.eda.plot_scatterXY(x, y, stride=1, plot_XequalsY=False, ax=None, **plt_kwargs)[source]¶ Plot a XY scatter plot of two Series.
Parameters: x, y : Pandas.Series
stride : Positive integer
If stride == n, then plot only every nth point
plot_XequalsY : Boolean
If True, plot the line X = Y in red.
plt_kwargs : Additional kwargs to pass to plt.scatter
-
rosetta.modeling.eda.reducedY_vs_binnedX(x, y, Y_reducer=<function mean>, X_reducer='midpoint', bins=10, quantiles=False, labels=None)[source]¶ Bin X and, inside every bin, apply Y_reducer to the Y values.
Parameters: x : Pandas.Series with numeric data
y : Pandas.Series with numeric data = 0 or 1
Y_reducer : function
Used to aggregate the Y values in every bin
X_reducer : function or ‘midpoint’
Used to aggregate the X values in every bin. This gives us the bin labels that are used as the indices. If ‘midpoint’, then use the bin midpoint.
bins : Positive Integer, optional
Number of bins to divide series
quantiles : Boolean, optional
If True, bin data using quantiles rather than an evenly divided range
labels : List-like, with len(labels) = len(x), optional
If given, use these labels to bin X rather than bins.
Returns: y_reduced : Series
The reduced y values with an index equal to the reduced X
count_X : Series
The number of X variables in each bin. Index is the reduced value.
Examples
Suppose Y is binary. Then to compute P[Y=1|X=x] (for x inside the bins), as well as #[X=x], use:
P_Y_g_X, count_X = eda.reducedY_vs_binnedX(x, y, Y_reducer=np.mean)
prediction_plotter¶
Helpful module for plotting predictions
-
class
rosetta.modeling.prediction_plotter.BasePlotter2D[source]¶ Abstract base class for 2D plotters. Not to be used directly.
Methods
-
plot(clf, X, y, mode='predict', contourf_kwargs={}, scatter_kwargs={})[source]¶ Plot levelsets of clf then plot the X/y data.
Parameters: clf : Trained sklearn classifier
mode : ‘predict’, ‘predict_proba’
If ‘predict’, plot the 0/1 levelsets using clf.predict If ‘predct_proba’, plot a contour plot of clf.predict_proba.
contourf_kwargs : Dict
kwargs passed to pylab.contourf
scatter_kwargs : Dict
kwargs passed to pylab.scatter
-
plot_levelsets(clf, box_ends=None, mode='predict', **contourf_kwargs)[source]¶ Plot level sets of the model clf.
Parameters: clf : Trained sklearn model
box_ends : 4-tuple
xmin, xmax, ymin, ymax plot levelsets within box defined by box_ends
box_ends : 4-tuple
xmin, xmax, ymin, ymax plot levelsets within box defined by box_ends Over-rides self.box_ends if self.box_ends is not set.
mode : ‘predict’, ‘predict_proba’
If ‘predict’, plot the levelsets using clf.predict If ‘predct_proba’, plot a contour plot of clf.predict_proba.
contourf_kwargs : Keyword arguments
Passed to pylab.contourf
-
-
class
rosetta.modeling.prediction_plotter.ClassifierPlotter2D(y_markers=None, y_names=None, x_names=None, cmap='PuBu', box_ends=None)[source]¶ For plotting 2D classifiers.
Initialize the ClassifierPlotter2D, then train different (2-d) classifiers on different data sets and plot the data and level sets of the classifier.
Methods
-
class
rosetta.modeling.prediction_plotter.RegressorPlotter2D(x_names=None, y_name=None, cmap='PuBu', box_ends=None)[source]¶ For plotting 2D regressors.
Initialize the RegressorPlotter2D, then train different (2-d) regressors on different data sets and plot the data and level sets of the regressor.
Methods
var_create¶
-
rosetta.modeling.var_create.build_xy_for_linearize(x, y, bins=10, Y_reducer=<function mean>, x_lims=None, endpoints=None)[source]¶ Return x and y for use in linearization. Use with var_create.interp.
Parameters: x : Pandas.Series
y : Pandas.Series
bins : positive integer
Number of bins for x
Y_reducer : Function
Used to reduce Y in each of the bins. E.g np.mean, logit_of_mean.
x_lims : 2-tuple, (xmin, xmax)
Rescaled x will be constant outside of this range. Choose xmin, xmax such that you have enough data in the interval (xmin, xmax)
endpoints : Array-like
[xmin, xmax, ymin, ymax]. Makes sure F(xmin) = ymin, etc...
Returns: x : Array
The bin midpoints (with adjunstments at the ends)
y : Array
y reduced in the bins
-
rosetta.modeling.var_create.interp(x, y, t=1, scaling=None)[source]¶ Return interpolation helpers for x and y. See build_xy_for_linearize for use in linearization.
Parameters: x : Array-like
y : Array-like
t : Real number in [0, 1]
With F(x) the linearization function, re-set F(x) = t*F(x) + (1-t)*x
scaling : String
- If None, the output is not rescaled and Y_reducer(bin_j) = x_j where
x_j is the midpoint of bin_j.
If ‘standardize’, then output will have zero mean and unit variance If ‘unit’, then output will be on the interval [0, 1]
Returns: F_x : Pandas.Series
A rescaled version of x.
F : Function that will rescale x
Cannot be pickled... :(
Examples
x4linear, y4linear = vc.build_xy_for_linearize(y_score, y) F_x, F = interp(x4linear, y4linear)
fitting¶
Common functions for fitting regression and classification models.
-
class
rosetta.modeling.fitting.CoefficientConverter(df, ones_column=None, dont_standardize=[], dont_winsorize=[], lower_quantile=0, upper_quantile=1, max_std=inf)[source]¶ For [un]standardizing/winsorizing coefficients and data. CoefficientConverter is initialized with one dataset, from this the standardization/winsorization rules are learned. The functions can be applied to other datasets.
- Standardization part of module provides the fundamental relation:
- X.dot(self.unstandardize_params(w_st)) = self.standardize(X).dot(w_st)
WORKFLOW 1 1) Initialize with a DataFrame. From this frame we learn the rules. 2) To fit, we use self.transform to transform a (possibly) new DataFrame.
This fit results in a set of “transformed params” w_tr- To predict Y_hat corresponding to new input X, we first compute X_tr = self.transform(X), and then use X_tr.dot(w_tr)
WORKFLOW 2 (standardization only!!) 1) Initialize with a dataframe. From this frame we learn the
standardization rules.- 2a) To fit, we use self.standardize to standardize a (possibly) new
- DataFrame. This fit results in a set of “standardized params” w_st.
- 2b) We obtain the “unstandardized params”
- w = self.unstandardized_params(w_st)
- To predict Y_hat corresponding to new input X, we use X.dot(w)
Methods
-
standardize(data)[source]¶ Returns a standardized version of data.
Parameters: data : pandas Series or DataFrame Notes
data is standardized according to the rules that self was initialized with, i.e. the rules implicit in self.stats.
-
rosetta.modeling.fitting.get_relative_error(reality, estimate)[source]¶ Compares estimate to reality and returns the the mean-square error:
|estimate - reality|_F / |reality|_F where F is the Frobenius norm.
-
rosetta.modeling.fitting.standardize(df, dont_standardize=None)[source]¶ Parameters: df : pandas DataFrame
Contains independent variables
dont_standardize : List
Names of variables to not standardize
Returns: Tuple of DataFrames: standardized_df, stats_df :
standardized_df is the standardized version of df stats_df contains the mean and std of the variables
-
rosetta.modeling.fitting.winsorize(series, lower_quantile=0, upper_quantile=1, max_std=inf)[source]¶ Truncate all items in series that are in extreme quantiles.
Parameters: series : pandas.Series. Real valued.
upper_quantile : Real number in [0, 1]
The upper quantile above which we trim
lower_quantile : Real number in [0, 1]
The lower quantile below which we trim
max_std : Non-negative real
Trim values that are more than max_std standard deviations away from the mean
Returns: winsorized_series : pandas.Series
Notes
Trimming according to max_std is done AFTER quantile trimming. I.e. the std is computed on the series that has already been trimmed by quantile.
categorical_fitter¶
Various functions for fitting categorical models. Put functions specific to logistic regression in multinomial_fitter
-
rosetta.modeling.categorical_fitter.predict_proba_cv(clf, X, y, n_folds=5)[source]¶ Returns an out-of-sample clf.predict_proba(X, y).
Parameters: clf : sklearn classifier with a predict_proba method
X : 2-D numpy array or DataFrame
y : 1-D numpy array or Series
Use this along with StratifiedKFold to determine splits.
n_folds: int :
Returns: probas : np.ndarray or series
Examples¶
modeling examples¶
prediction_plotter examples¶
"""
An example script plotting some classifiers using prediction_plotter.
"""
import numpy as np
import matplotlib.pylab as pl
pl.ion()
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
from sklearn import svm
from sklearn.neighbors import KNeighborsClassifier
from rosetta.modeling import prediction_plotter
###############################################################################
# Make training data
###############################################################################
# X data
N = 50 # Number of data points
x1min, x1max, x2min, x2max = 0., 200., 1., 20.
x1 = np.random.randint(x1min, x1max+1, size=N)
x2 = np.random.randint(x2min, x2max+1, size=N)
X = np.c_[x1, x2]
# y data
# Probability y = 1 is highest near the mid-points of X
x1mid = (x1max - x1min) / 2.
x2mid = (x2max - x2min) / 2.
center = np.array([x1mid, x2mid])
width1, width2 = x1mid/2, x2mid/2
product = (
((X[:, 0] - center[0]) / width1)**4 + ((X[:, 1] - center[1]) / width2)**4)
pdf_arr = np.exp(- product / 2. )
y = (np.random.rand(N) < pdf_arr).astype('int')
# Names
x_names = ['doc-length', 'num-recipients']
y_names = ['non-relevant', 'relevant']
y_markers = ['x', 'o']
###############################################################################
# Initialize the plotter
###############################################################################
plotter = prediction_plotter.ClassifierPlotter2D(
y_markers=y_markers, y_names=y_names, x_names=x_names)
###############################################################################
# Decision Tree
###############################################################################
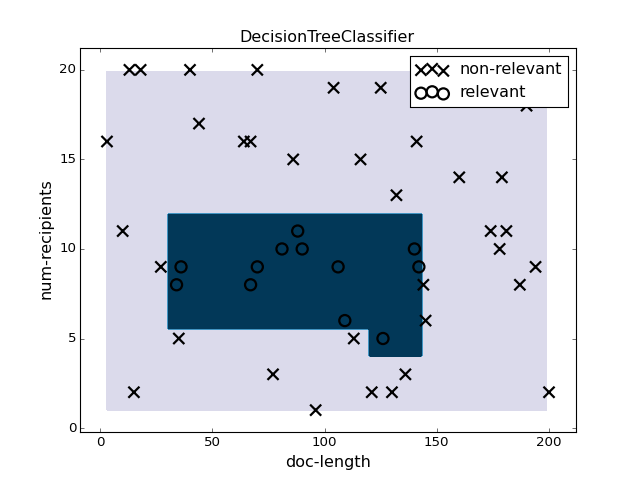
pl.figure(1)
pl.clf()
clf = DecisionTreeClassifier().fit(X, y)
plotter.plot(clf, X, y)
pl.title("DecisionTreeClassifier")
###############################################################################
# Random Forest
###############################################################################
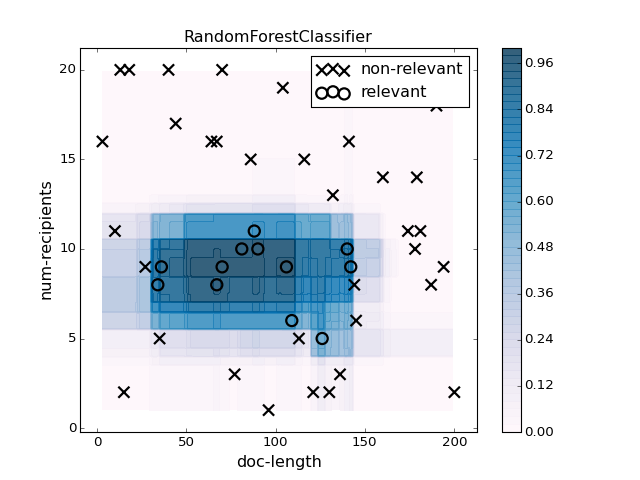
pl.figure(2)
pl.clf()
clf = RandomForestClassifier(n_estimators=100, max_features=None).fit(X, y)
plotter.plot(clf, X, y, mode='predict_proba', contourf_kwargs={'alpha':0.8})
pl.title("RandomForestClassifier")
###############################################################################
# Logistic Regression
###############################################################################
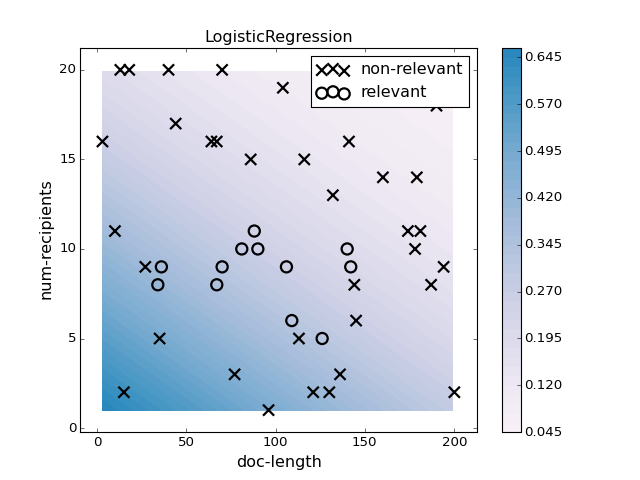
pl.figure(3)
pl.clf()
clf = LogisticRegression(penalty='l2', C=1000).fit(X, y)
plotter.plot(clf, X, y, mode='predict_proba')
pl.title("LogisticRegression")
###############################################################################
# SVM
###############################################################################
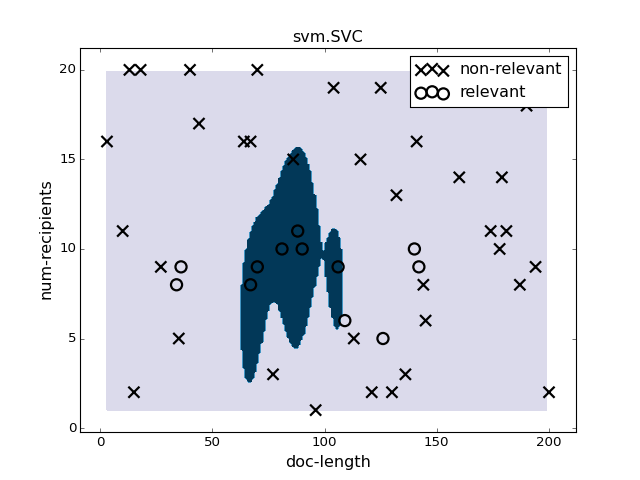
pl.figure(4)
pl.clf()
clf = svm.SVC(gamma=0.01).fit(X, y)
plotter.plot(clf, X, y)
pl.title("svm.SVC")
###############################################################################
# K Nearest Neighbors
###############################################################################
pl.figure(5)
pl.clf()
clf = KNeighborsClassifier(n_neighbors=N/10).fit(X, y)
plotter.plot(clf, X, y)
pl.title("KNeighborsClassifier")
"""
An example script plotting some regressors using prediction_plotter.
"""
import numpy as np
import matplotlib.pylab as pl
pl.ion()
from sklearn.linear_model import LinearRegression
from sklearn.neighbors import KNeighborsRegressor
from sklearn import svm
from sklearn.gaussian_process import GaussianProcess
from rosetta.modeling import prediction_plotter
###############################################################################
# Make training data
###############################################################################
# X data
N = 10 # Number of data points
x1min, x1max, x2min, x2max = 0., 200., 1., 20.
x1 = np.random.randint(x1min, x1max+1, size=N)
x2 = np.random.randint(x2min, x2max+1, size=N)
X = np.c_[x1, x2]
# y data
# y is bigger near the center of the support of X
# noise is added to y
x1mid = (x1max - x1min) / 2.
x2mid = (x2max - x2min) / 2.
center = np.array([x1mid, x2mid])
width1, width2 = x1mid/2, x2mid/2
noise_level = 0.2
product = ((X[:, 0] - center[0]) / width1)**4 + ((X[:, 1] - center[1]) / width2)**4
y = np.exp(- product / 2. ) + noise_level * np.random.randn(N)
###############################################################################
# Initialize the plotter
###############################################################################
plotter = prediction_plotter.RegressorPlotter2D(
x_names=['age', 'height'], y_name='measured-data')
###############################################################################
# Linear Regression
###############################################################################
pl.figure(1)
pl.clf()
clf = LinearRegression().fit(X, y)
plotter.plot(clf, X, y)
pl.title("LinearRegression")
pl.colorbar()
###############################################################################
# K Nearest Neighbors
###############################################################################
pl.figure(2)
pl.clf()
n_neighbors = max(2, N/10)
clf = KNeighborsRegressor(n_neighbors=n_neighbors).fit(X, y)
plotter.plot(clf, X, y)
pl.title("KNeighborsRegressor")
pl.colorbar()
###############################################################################
# Support Vector Regressor (SVR, a.k.a. RVM)
###############################################################################
pl.figure(4)
pl.clf()
clf = svm.SVR(gamma=0.01).fit(X, y)
plotter.plot(clf, X, y)
pl.title("svm.SVR")
pl.colorbar()
###############################################################################
# Gaussian Process Models
###############################################################################
pl.figure(5)
pl.clf()
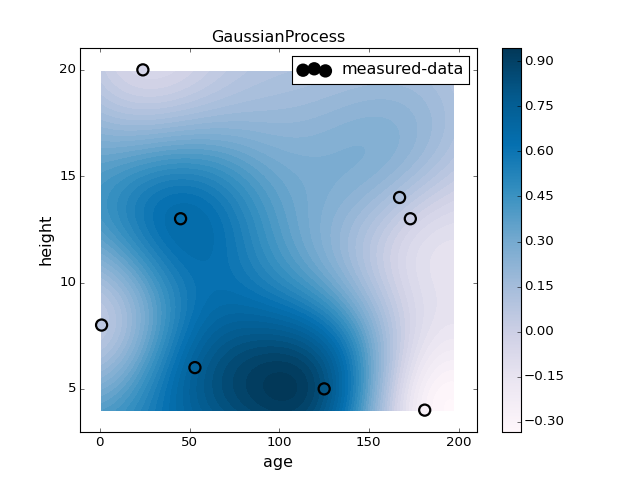
clf = GaussianProcess(regr='linear', theta0=2).fit(X, y)
plotter.plot(clf, X, y)
pl.title("GaussianProcess")
pl.colorbar()
eda examples¶
"""
Examples using rosetta.modeling.eda
"""
import pandas as pd
import numpy as np
import matplotlib.pylab as pl
from numpy.random import randn, rand
from rosetta.modeling import eda
###############################################################################
# X-Y plotting
###############################################################################
N = 1000
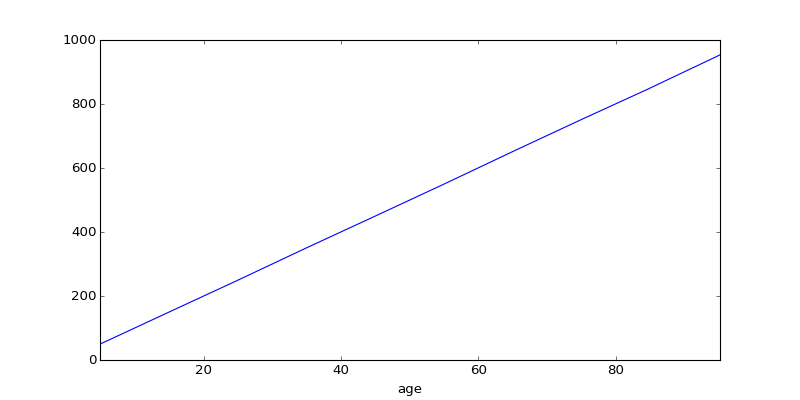
# Make a linear income vs. age relationship.
age = pd.Series(100 * rand(N))
age.name = 'age'
income = 10 * age + 10 * randn(N)
income.name = 'income'
# The relationship E[Y | X=x] is linear
pl.figure(1); pl.clf()
eda.plot_reducedY_vs_binnedX(age, income)
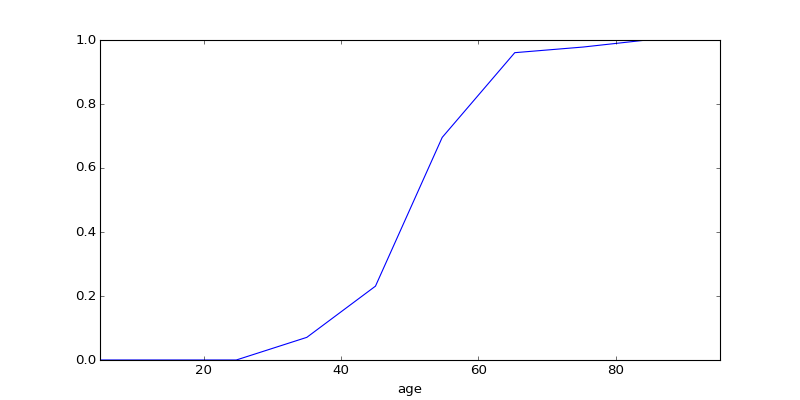
# Make a sigmoidal P[is_manager | X=x] relationship
def sigmoid(x):
x_st = 5 * (x - x.mean()) / x.std()
return np.exp(x_st) / (1 + np.exp(x_st))
is_manager = (rand(N) < sigmoid(age)).astype('int')
is_manager.name = 'is_manager'
pl.figure(2); pl.clf()
eda.plot_reducedY_vs_binnedX(age, is_manager)
###############################################################################
# Correlation matrix plotting
###############################################################################
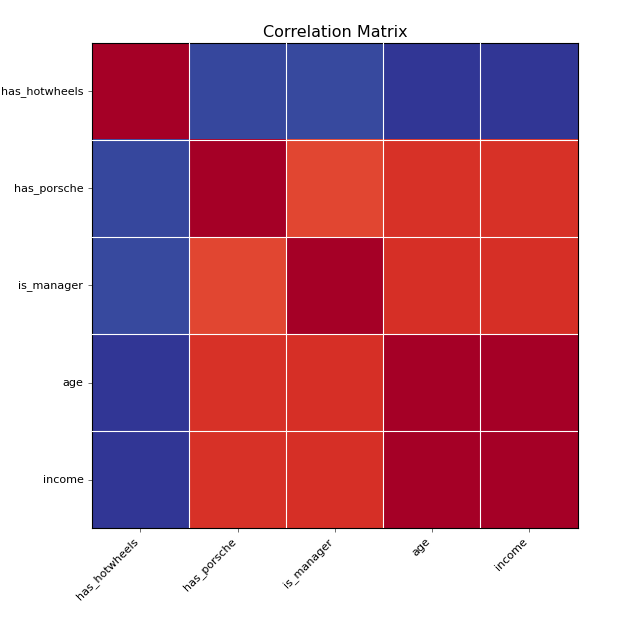
# P[has_porsche] is higher for young rich people
has_porsche = (rand(N) < sigmoid(income - 0.5 * age)).astype('int')
has_porsche.name = 'has_porsche'
has_hotwheels = (rand(N) < sigmoid(-age)).astype('int')
has_hotwheels.name = 'has_hotwheels'
all_vars = pd.concat(
[age, income, is_manager, has_porsche, has_hotwheels], axis=1)
corr = all_vars.corr()
fig = pl.figure(3); pl.clf()
eda.plot_corr_grid(corr)
fig = pl.figure(4); pl.clf()
eda.plot_corr_dendrogram(corr)