1. Quick Start¶
Contents
In order to use any of the CNO formalisms, you will need
- a network of proteins also known as PKN (for prior knowledge network) or model
- a data file that contains phosphorylation data sets (perturbations) in a MIDAS format
- decide on a formalism
1.1. Conventions¶
We are going to manipulate protein networks but from a logical perspective. So, let us give the conventions being used to encode the relations between proteins.
The reactions are encoded using the = sign with inputs on the left hand side (LHS) and output on the RHS. There is only one output but possibly 1 or more inputs. If several inputs are provided, the logic could be OR or AND or a mix of them. OR relations are encoded with + sign and AND relations are encoded with the ^ sign. Those relations are correct:
A=B
A+B=C
A^C=D
E+F^G=H
The class Reaction can help you to validate a reaction.:
from cno import Reaction
r = Reaction("A+B=C")
Activation are encoded as above. Inhibition are encode using the ! character.:
!A=B
means A inhibits B.
See also
See cno.io.reactions for more details
1.2. Your input model (PKN model)¶
The input data file should be encoded using the SIF format but SBML-qual format are also accepted using the SIF or SBMLqual classes.
The SIF format is a 3-column tab separated value format. The parsing is flexible enough that any white spaces are considered as the separator. The LHS column contain the input species and the RHS column contains the output species. The middle column contains the type of relation:
- -1 is inhbition
- 1 is activation
In the following we will use a more advanced data structure called CNOGraph, which will ease the manipulations of both the protein network and data set (MIDAS). Some examples of PKN models and data sets are provided in the cno.datasets package. A convenient function to retrieve filename and path of those examples is called cnodata():
from cno import cnodata
filename = cnodata("PKN-ToyMMB.sif")
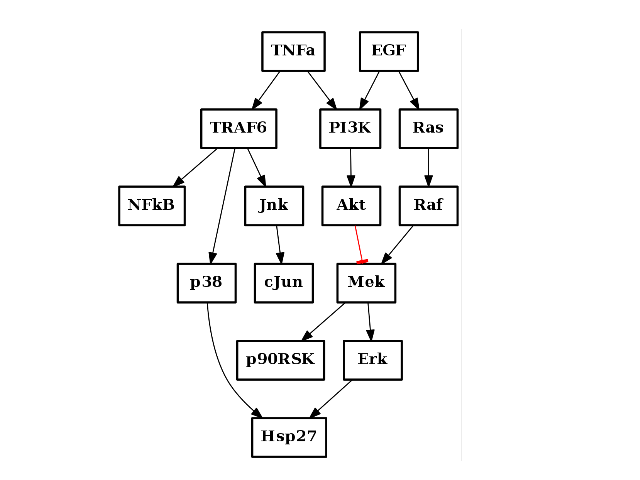
The **CNOGraph** structure can read SIF file (or SBMLqual). As an example,
we fetch a local filename and plot its graphical representation:
from cno import CNOGraph, SIF, cnodata
filename = cnodata("PKN-ToyMMB.sif")
c = CNOGraph(filename)
c.plot()
(Source code, png, hires.png, pdf)
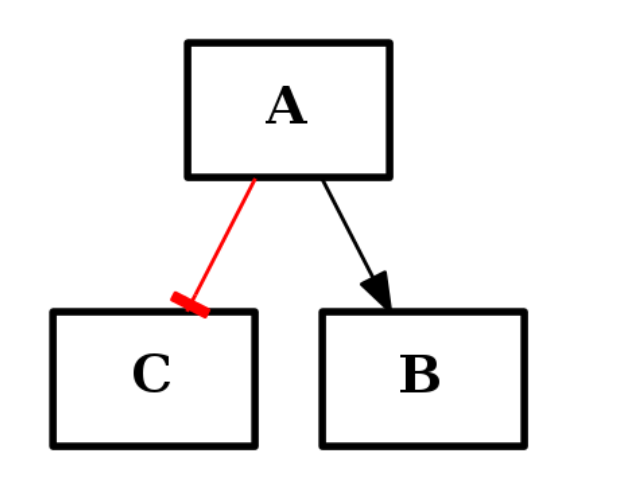
The CNOGraph is a DiGraph data structure, which can also be built from scratch and re-used in other context. There is a current restriction though, which is that edge type have to be provided as on the type of edges that can be only of two types: “+” for activation and “-” for inhibition. The following example shows how to create a simple graph made of 3 nodes and 2 edges: one activation (black) and one inhibition (red):
from cno import CNOGraph
c1 = CNOGraph()
c1.add_edge("A","B", link="+")
c1.add_edge("A","C", link="-")
c1.plot()
(Source code, png, hires.png, pdf)
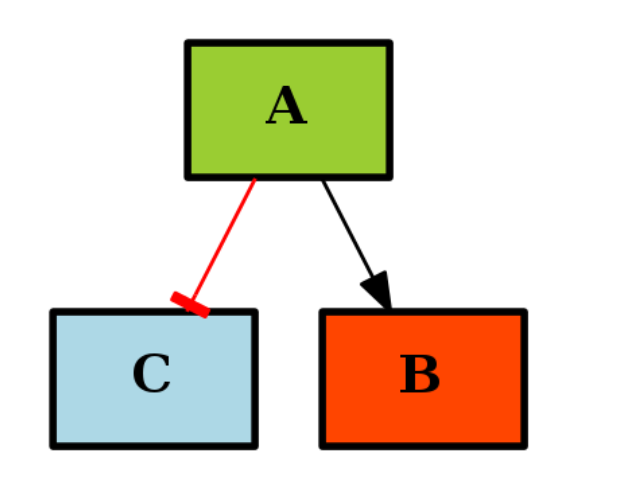
If you use a MIDAS file during the instanciation, the CNOGraph will color the nodes that are found in the MIDAS file. Stimuli (ligand) are colored in green, inhibitors in red and readout (signal) in blue. If you did not provide a MIDAS file, you can still specificy the list manually like in the following example:
from cno import CNOGraph
c1 = CNOGraph()
c1.add_edge("A","B", link="+")
c1.add_edge("A","C", link="-")
c1._stimuli = ["A"]
c1._inhibitors = ["B"]
c1._signals = ["C"]
c1.plot()
c1.to_sif("test.sif")
(Source code, png, hires.png, pdf)
There are many operators available and readers can refer to cno.io.cnograph.CNOGraph for more examples.
1.3. The input data set (MIDAS)¶
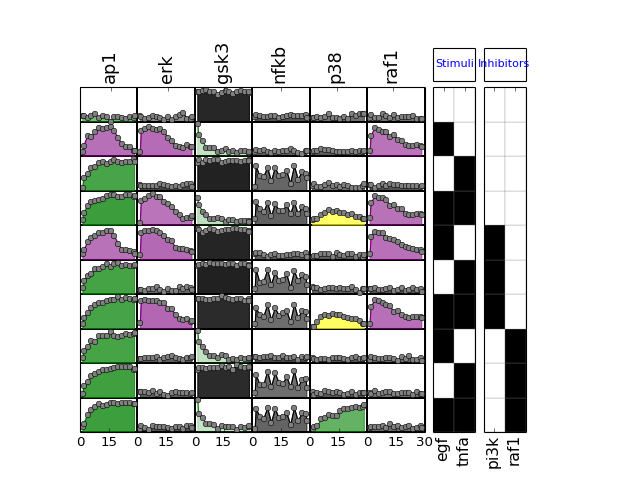
The MIDAS data file can be read using the XMIDAS class, which contains a few methods described in other sections or notebooks.
from cno import XMIDAS, cnodata
m = XMIDAS(cnodata("MD-ToyPB.csv"))
m.plot()
(Source code, png, hires.png, pdf)
1.4. Boolean formalism example¶
The goal of CNO is to provide a set of tools to optimise PKN to data sets using various logical formalism. The optimisation and logical simulations are currently perfomred using CellNOptR. The formalisms available are
- steady state using boolean approach
- discrete time using boolean asynchronous approach
- logical ode formalism
- fuzzy approach using Hill function on the edges but using boolean approach for logical gates (min and max of the inputs)
Here below we show the first case. All other formalism would have similar user interface.
from cno import CNORbool, cnodata
c = CNORbool(cnodata("PKN-ToyMMB.sif"), cnodata("MD-ToyMMB.csv"))
c.optimise()
c.plot_fitness()
c.plot_errors()
c.results.results.best_bitstring()
c.results.results.best_score()
# open a report page in a browser
# c.onweb()