Application Example of GP regression¶
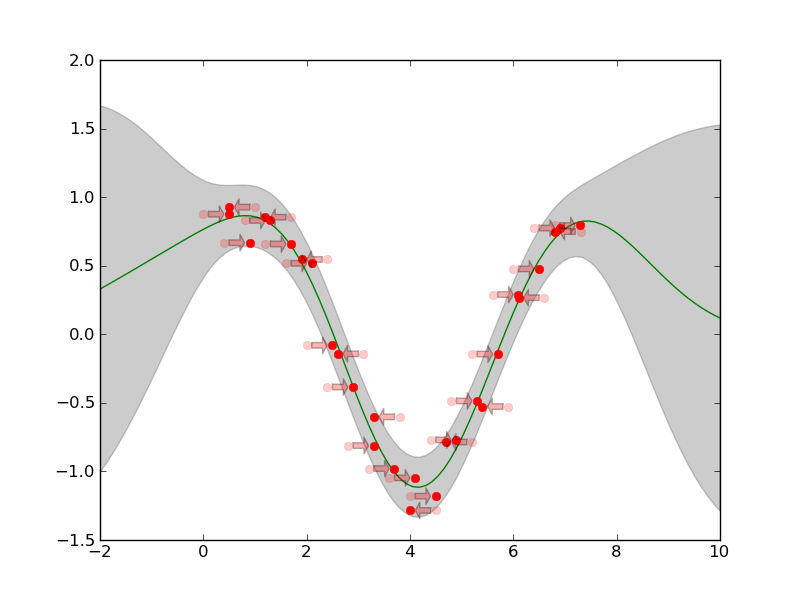
This Example shows the Squared Exponential CF (pygp.covar.se.SEARDCF) preprocessed by shiftCF(:py:class`covar.combinators.ShiftCF) and combined with noise pygp.covar.noise.NoiseISOCF by summing them up (using pygp.covar.combinators.SumCF). We will shift two input replicates against each other, to make them fit to each other.
First of all we have to import all important packages:
import pylab as PL
import scipy as SP
import numpy.random as random
Now import the Covariance Functions and Combinators:
from pygp.covar import se, noise, combinators
And additionally the GP regression framework (pygp.gp, pygp.priors for the priors and pygp.plot.gpr_plot for plotting the results):
from pygp.gp.basic_gp import GP
from pygp.optimize.optimize import opt_hyper
from pygp.priors import lnpriors
import pygp.plot.gpr_plot as gpr_plot
For this particular example we generate some simulated random sinus data; just samples from a superposition of a sin + linear trend:
xmin = 1
xmax = 2.5*SP.pi
x1 = SP.arange(xmin,xmax,.7)
x2 = SP.arange(xmin,xmax,.4)
C = 2 #offset
b = 0.5
sigma = 0.1
b = 0
y1 = b*x1 + C + 1*SP.sin(x1)
dy1 = b + 1*SP.cos(x1)
y1 += sigma*random.randn(y1.shape[0])
y1-= y1.mean()
y2 = b*x2 + C + 1*SP.sin(x2)
dy2 = b + 1*SP.cos(x2)
y2 += sigma*random.randn(y2.shape[0])
y2-= y2.mean()
x1 = x1[:,SP.newaxis]
x2 = (x2-1)[:,SP.newaxis]
x = SP.concatenate((x1,x2),axis=0)
y = SP.concatenate((y1,y2),axis=0)
The predictions we will make on the interpolation interval
X = SP.linspace(-2,10,100)[:,SP.newaxis]
For the calculation of the replicates, we need to give the replicate indices per input x:
dim = 1
replicate_indices = SP.concatenate([SP.repeat(i,len(xi)) for i,xi in enumerate((x1,x2))])
n_replicates = len(SP.unique(replicate_indices))
Thus, our starting hyperparameters are:
logthetaCOVAR = SP.log([1,1,1,1,sigma])
hyperparams = {'covar':logthetaCOVAR}
Now the interesting point: creating the sumCF by combining noise and se:
SECF = se.SEARDCF(dim)
noiseCF = noise.NoiseISOCF()
shiftCF = combinators.ShiftCF(SECF,replicate_indices)
covar = combinators.SumCF((shiftCF,noiseCF))
And the prior believes, we have about the hyperparameters:
covar_priors = []
#Length-Scale
covar_priors.append([lnpriors.lngammapdf,[1,2]])
for i in range(dim):
covar_priors.append([lnpriors.lngammapdf,[1,1]])
#X-Shift
for i in range(n_replicates):
covar_priors.append([lnpriors.lngausspdf,[0,.5]])
#Noise
covar_priors.append([lnpriors.lngammapdf,[1,1]])
priors = {'covar':covar_priors}
We want all hyperparameters to be optimized:
Ifilter = {'covar': SP.array([1,1,1,1,1],dtype='int')}
Create the GP regression class for further usage:
gpr = GP(covar,x=x,y=y)
And optimize the hyperparameters:
[opt_model_params,opt_lml]=opt_hyper(gpr,hyperparams,priors=priors,gradcheck=False,Ifilter=Ifilter)
With these optimized hyperparameters we can now predict the point-wise mean M and deviance S of the training data:
[M,S] = gpr.predict(opt_model_params,X)
For the sake of beauty plot the mean M and deviance S:
T = opt_model_params['covar'][2:4]
gpr_plot.plot_sausage(X,M,SP.sqrt(S))
gpr_plot.plot_training_data(x,y,shift=T,replicate_indices=replicate_indices)
The resulting plot is: