Doctests¶
-
skfmm.testing()[source]¶ These tests are gathered from FiPy, PyLSMLIB and original Scikit-fmm tests.
1D Test
>>> import numpy as np
>>> print(np.allclose(distance((-1., -1., -1., -1., 1., 1., 1., 1.), dx=.5), ... (-1.75, -1.25, -.75, -0.25, 0.25, 0.75, 1.25, 1.75))) True
Small dimensions.
>>> dx = 1e-10 >>> print(np.allclose(distance((-1., -1., -1., -1., 1., 1., 1., 1.), dx=dx), ... np.arange(8) * dx - 3.5 * dx)) True
Bug Fix
Test case for a bug in the upwind finite difference scheme for negative phi. When computing finite differences we want to preferentially use information from the frozen neighbors that are closest to the zero contour in each dimension. This means that we must compare absolute distances when checking neighbors in the negative phi direction.
The error can result in incorrect values of the updated signed distance function for regions close to the minimum contour of the level set function, i.e. in the middle of holes.
To test we use a square matrix for the initial phi field that is equal to -1 on the main diagonal and on the three diagonals above and below this. The matrix is set to 1 everywhere else. The bug results in errors in positions (1,1), (2,2), (3,3), (6,6), (7,7) and (8,8) along the main diagonal.
This error occurs for first- and second-order updates. For simplicity, we choose to only test the first-order update.
>>> phi = np.ones((10, 10)) >>> i,j = np.indices(phi.shape) >>> phi[i==j-3] = -1 >>> phi[i==j-2] = -1 >>> phi[i==j-1] = -1 >>> phi[i==j] = -1 >>> phi[i==j+1] = -1 >>> phi[i==j+2] = -1 >>> phi[i==j+3] = -1 >>> phi = distance(phi, order=1) >>> print(np.allclose(phi[1, 1], -2.70464, atol=1e-4)) True >>> print(np.allclose(phi[2, 2], -2.50873, atol=1e-4)) True >>> print(np.allclose(phi[3, 3], -2.47487, atol=1e-4)) True >>> print(np.allclose(phi[6, 6], -2.47487, atol=1e-4)) True >>> print(np.allclose(phi[7, 7], -2.50873, atol=1e-4)) True >>> print(np.allclose(phi[8, 8], -2.70464, atol=1e-4)) True
Bug Fix
A 2D test case to test trial values for a pathological case.
>>> dx = 1. >>> dy = 2. >>> vbl = -dx * dy / np.sqrt(dx**2 + dy**2) / 2. >>> vbr = dx / 2 >>> vml = dy / 2. >>> crossProd = dx * dy >>> dsq = dx**2 + dy**2 >>> top = vbr * dx**2 + vml * dy**2 >>> sqrt = crossProd**2 *(dsq - (vbr - vml)**2) >>> sqrt = np.sqrt(max(sqrt, 0)) >>> vmr = (top + sqrt) / dsq >>> print(np.allclose(distance(((-1., 1., -1.), (1., 1., 1.)), dx=(dx, dy), order=1), ... ((vbl, vml, vbl), (vbr, vmr, vbr)))) True
Test Extension Field Calculation
>>> tmp = 1 / np.sqrt(2) >>> phi = np.array([[-1., 1.], [1., 1.]]) >>> phi, ext = extension_velocities(phi, ... [[-1, .5], [2., -1.]], ... ext_mask=phi < 0, ... dx=1., order=1) >>> print(np.allclose(phi, ((-tmp / 2, 0.5), (0.5, 0.5 + tmp)))) True >>> print(np.allclose(ext, [[1.25, .5], [2., 1.25]])) True
>>> phi = np.array(((-1., 1., 1.), (1., 1., 1.), (1., 1., 1.))) >>> phi, ext = extension_velocities(phi, ... ((-1., 2., -1.), ... (.5, -1., -1.), ... (-1., -1., -1.)), ... ext_mask=phi < 0, ... order=1) >>> v1 = 0.5 + tmp >>> v2 = 1.5 >>> tmp1 = (v1 + v2) / 2 + np.sqrt(2. - (v1 - v2)**2) / 2 >>> tmp2 = tmp1 + 1 / np.sqrt(2) >>> print(np.allclose(phi, ((-tmp / 2, 0.5, 1.5), ... (0.5, 0.5 + tmp, tmp1), ... (1.5, tmp1, tmp2)))) True >>> print(np.allclose(ext, ((1.25, 2., 2.), ... (.5, 1.25, 1.5456), ... (.5, 0.9544, 1.25)), ... rtol = 1e-4)) True
Bug Fix
Test case for a bug that occurs when initializing the distance variable at the interface. Currently it is assumed that adjacent cells that are opposite sign neighbors have perpendicular normal vectors. In fact the two closest cells could have opposite normals.
>>> print(np.allclose(distance((-1., 1., -1.)), (-0.5, 0.5, -0.5))) True
Testing second order. This example failed with Scikit-fmm.
>>> phi = ((-1., -1., 1., 1.), ... (-1., -1., 1., 1.), ... (1., 1., 1., 1.), ... (1., 1., 1., 1.)) >>> answer = ((-1.30473785, -0.5, 0.5, 1.49923009), ... (-0.5, -0.35355339, 0.5, 1.45118446), ... (0.5, 0.5, 0.97140452, 1.76215286), ... (1.49923009, 1.45118446, 1.76215286, 2.33721352)) >>> print(np.allclose(distance(phi), ... answer, ... rtol=1e-9)) True
A test for a bug in both LSMLIB and Scikit-fmm
The following test gives different results depending on whether LSMLIB or Scikit-fmm is used. This issue occurs when calculating second order accurate distance functions. When a value becomes “known” after previously being a “trial” value it updates its neighbors’ values. In a second order scheme the neighbors one step away also need to be updated (if the cell between the new “known” cell and the cell required for second order accuracy also happens to be “known”), but are not updated in either package. By luck (due to trial values having the same value), the values calculated in Scikit-fmm for the following example are correct although an example that didn’t work for Scikit-fmm could also be constructed.
>>> phi = distance([[-1, -1, -1, -1], ... [ 1, 1, -1, -1], ... [ 1, 1, -1, -1], ... [ 1, 1, -1, -1]], order=2) >>> phi = distance(phi, order=2)
The following values come form Scikit-fmm.
>>> answer = [[-0.5, -0.58578644, -1.08578644, -1.85136395], ... [ 0.5, 0.29289322, -0.58578644, -1.54389939], ... [ 1.30473785, 0.5, -0.5, -1.5 ], ... [ 1.49547948, 0.5, -0.5, -1.5 ]]
The 3rd and 7th element are different for LSMLIB. This is because the 15th element is not “known” when the “trial” value for the 7th element is calculated. Scikit-fmm calculates the values in a slightly different order so gets a seemingly better answer, but this is just chance.
>>> print(np.allclose(phi, answer, rtol=1e-9)) True
The following tests for the same issue but is a better test case guaranteed to fail.
>>> phi = np.array([[-1, 1, 1, 1, 1, -1], ... [-1, -1, -1, -1, -1, -1], ... [-1, -1, -1, -1, -1, -1]])
>>> phi = distance(phi) >>> print(phi[2, 2] == phi[2, 3]) True
>>> phi = distance(phi) >>> print(phi[2, 2] == phi[2, 3]) True
Circle Example
Solve the level set equation in two dimensions for a circle.
The 2D level set equation can be written,
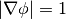
and the boundary condition for a circle is given by,
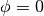 at
at
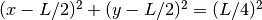 .
.The solution to this problem will be demonstrated in the following script. Firstly, setup the parameters.
>>> def mesh(nx=1, ny=1, dx=1., dy=1.): ... y, x = np.mgrid[0:nx,0:ny] ... x = x * dx + dx / 2 ... y = y * dy + dy / 2 ... return x, y
>>> dx = 1. >>> N = 11 >>> L = N * dx >>> x, y = mesh(nx=N, ny=N, dx=dx, dy=dx) >>> phi = -np.ones(N * N, 'd') >>> phi[(x.flatten() - L / 2.)**2 + (y.flatten() - L / 2.)**2 < (L / 4.)**2] = 1. >>> phi = np.reshape(phi, (N, N)) >>> phi = distance(phi, dx=dx, order=1).flatten()
>>> dX = dx / 2. >>> m1 = dX * dX / np.sqrt(dX**2 + dX**2) >>> def evalCell(phix, phiy, dx): ... aa = dx**2 + dx**2 ... bb = -2 * ( phix * dx**2 + phiy * dx**2) ... cc = dx**2 * phix**2 + dx**2 * phiy**2 - dx**2 * dx**2 ... sqr = np.sqrt(bb**2 - 4. * aa * cc) ... return ((-bb - sqr) / 2. / aa, (-bb + sqr) / 2. / aa) >>> v1 = evalCell(-dX, -m1, dx)[0] >>> v2 = evalCell(-m1, -dX, dx)[0] >>> v3 = evalCell(m1, m1, dx)[1] >>> v4 = evalCell(v3, dX, dx)[1] >>> v5 = evalCell(dX, v3, dx)[1] >>> MASK = -1000. >>> trialValues = np.array(( ... MASK, MASK, MASK, MASK, MASK, MASK, MASK, MASK, MASK, MASK, MASK, ... MASK, MASK, MASK, MASK,-3*dX,-3*dX,-3*dX, MASK, MASK, MASK, MASK, ... MASK, MASK, MASK, v1, -dX, -dX, -dX, v1, MASK, MASK, MASK, ... MASK, MASK, v2, -m1, m1, dX, m1, -m1, v2, MASK, MASK, ... MASK, -dX*3, -dX, m1, v3, v4, v3, m1, -dX,-dX*3, MASK, ... MASK, -dX*3, -dX, dX, v5, MASK, v5, dX, -dX,-dX*3, MASK, ... MASK, -dX*3, -dX, m1, v3, v4, v3, m1, -dX,-dX*3, MASK, ... MASK, MASK, v2, -m1, m1, dX, m1, -m1, v2, MASK, MASK, ... MASK, MASK, MASK, v1, -dX, -dX, -dX, v1, MASK, MASK, MASK, ... MASK, MASK, MASK, MASK,-3*dX,-3*dX,-3*dX, MASK, MASK, MASK, MASK, ... MASK, MASK, MASK, MASK, MASK, MASK, MASK, MASK, MASK, MASK, MASK), 'd')
>>> phi[trialValues == MASK] = MASK >>> print(np.allclose(phi, trialValues)) True
Square Example
Here we solve the level set equation in two dimensions for a square. The equation is given by:
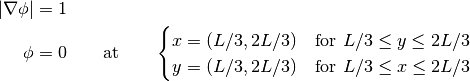
>>> dx = 0.5 >>> dy = 2. >>> nx = 5 >>> ny = 5 >>> Lx = nx * dx >>> Ly = ny * dy
>>> x, y = mesh(nx=nx, ny=ny, dx=dx, dy=dy) >>> x = x.flatten() >>> y = y.flatten() >>> phi = -np.ones(nx * ny, 'd') >>> phi[((Lx / 3. < x) & (x < 2. * Lx / 3.)) & ((Ly / 3. < y) & (y < 2. * Ly / 3))] = 1. >>> phi = np.reshape(phi, (nx, ny)) >>> phi = distance(phi, dx=(dy, dx), order=1).flatten()
>>> def evalCell(phix, phiy, dx, dy): ... aa = dy**2 + dx**2 ... bb = -2 * ( phix * dy**2 + phiy * dx**2) ... cc = dy**2 * phix**2 + dx**2 * phiy**2 - dx**2 * dy**2 ... sqr = np.sqrt(bb**2 - 4. * aa * cc) ... return ((-bb - sqr) / 2. / aa, (-bb + sqr) / 2. / aa) >>> val = evalCell(-dy / 2., -dx / 2., dx, dy)[0] >>> v1 = evalCell(val, -3. * dx / 2., dx, dy)[0] >>> v2 = evalCell(-3. * dy / 2., val, dx, dy)[0] >>> v3 = evalCell(v2, v1, dx, dy)[0] >>> v4 = dx * dy / np.sqrt(dx**2 + dy**2) / 2 >>> arr = np.array(( ... v3 , v2 , -3. * dy / 2. , v2 , v3, ... v1 , val , -dy / 2. , val , v1 , ... -3. * dx / 2., -dx / 2., v4 , -dx / 2., -3. * dx / 2., ... v1 , val , -dy / 2. , val , v1 , ... v3 , v2 , -3. * dy / 2. , v2 , v3 )) >>> print(np.allclose(arr, phi)) True
Assertion Errors
>>> distance([[-1, 1],[1, 1]], dx=(1, 2, 3)) Traceback (most recent call last): ... ValueError: dx must be of length len(phi.shape) >>> extension_velocities([[-1, 1],[1, 1]], speed=[1, 1]) Traceback (most recent call last): ... ValueError: phi and speed must have the same shape
Test for 1D equality between `distance` and `travel_time`
>>> phi = np.arange(-5, 5) + 0.499 >>> d = distance(phi) >>> t = travel_time(phi, speed=np.ones_like(phi)) >>> np.testing.assert_allclose(t, np.abs(d))
Tests taken from FiPy
>>> phi = np.array(((-1, -1, 1, 1), ... (-1, -1, 1, 1), ... (1, 1, 1, 1), ... (1, 1, 1, 1))) >>> o1 = distance(phi, order=1, self_test=True) >>> dw_o1 = [[-1.20710678, -0.5, 0.5, 1.5], ... [-0.5, -0.35355339, 0.5, 1.5], ... [ 0.5, 0.5, 1.20710678, 2.04532893], ... [ 1.5, 1.5, 2.04532893, 2.75243571]] >>> np.testing.assert_allclose(o1, dw_o1)
>>> phi = np.array(((-1, -1, 1, 1), ... (-1, -1, 1, 1), ... (1, 1, 1, 1), ... (1, 1, 1, 1))) >>> o1 = travel_time(phi, np.ones_like(phi), order=1, self_test=True) >>> dw_o1 = [[-1.20710678, -0.5, 0.5, 1.5], ... [-0.5, -0.35355339, 0.5, 1.5], ... [ 0.5, 0.5, 1.20710678, 2.04532893], ... [ 1.5, 1.5, 2.04532893, 2.75243571]] >>> np.testing.assert_allclose(o1, np.abs(dw_o1))
>>> phi = np.array(((-1, -1, 1, 1), ... (-1, -1, 1, 1), ... (1, 1, 1, 1), ... (1, 1, 1, 1))) >>> o2 = distance(phi, self_test=True) >>> dw_o2 = [[-1.30473785, -0.5, 0.5, 1.49923009], ... [-0.5, -0.35355339, 0.5, 1.45118446], ... [ 0.5, 0.5, 0.97140452, 1.76215286], ... [ 1.49923009, 1.45118446, 1.76215286, 2.33721352]]
>>> np.testing.assert_allclose(o2, dw_o2) >>> phi = np.array(((-1, -1, 1, 1), ... (-1, -1, 1, 1), ... (1, 1, 1, 1), ... (1, 1, 1, 1))) >>> o2 = travel_time(phi, np.ones_like(phi), self_test=True) >>> dw_o2 = [[-1.30473785, -0.5, 0.5, 1.49923009], ... [-0.5, -0.35355339, 0.5, 1.45118446], ... [ 0.5, 0.5, 0.97140452, 1.76215286], ... [ 1.49923009, 1.45118446, 1.76215286, 2.33721352]] >>> np.testing.assert_allclose(o2, np.abs(dw_o2))
>>> distance([-1,1], order=0) Traceback (most recent call last): ... ValueError: order must be 1 or 2
>>> distance([-1,1], order=3) Traceback (most recent call last): ... ValueError: order must be 1 or 2
Extension velocity tests
Test 1d extension constant.
>>> phi = [-1,-1,-1,1,1,1] >>> speed = [1,1,1,1,1,1] >>> d, f_ext = extension_velocities(phi, speed, self_test=True) >>> np.testing.assert_allclose(speed, f_ext)
Test the 1D extension block.
>>> phi = np.ones(10) >>> phi[0] =- 1 >>> speed = np.ones(10) >>> speed[0:3] = 5 >>> d, f_ext = extension_velocities(phi, speed, self_test=True) >>> np.testing.assert_allclose(f_ext, 5)
Test that a uniform speed value is preserved.
>>> N = 50 >>> X, Y = np.meshgrid(np.linspace(-1, 1, N), np.linspace(-1, 1, N)) >>> r = 0.25 >>> dx = 2.0 / (N - 1) >>> phi = (X) ** 2 + (Y) ** 2 - r ** 2 >>> speed = np.ones_like(phi) >>> d, f_ext = extension_velocities(phi, speed, dx, self_test=True) >>> np.testing.assert_allclose(f_ext, 1.0)
Constant value march-out test
>>> speed[abs(Y)<0.3] = 10.0 >>> d, f_ext = extension_velocities(phi, speed, dx, self_test=True) >>> np.testing.assert_allclose(f_ext, 10.0)
Test distance from extension
>>> speed = np.ones_like(phi) >>> d, f_ext = extension_velocities(phi, speed, dx, self_test=True) >>> d2 = distance(phi, dx, self_test=True) >>> np.testing.assert_allclose(d, d2)
Test for extension velocity bug
>>> N = 150 >>> X, Y = np.meshgrid(np.linspace(-1, 1, N), np.linspace(-1, 1, N)) >>> r = 0.5 >>> dx = 2.0 / (N - 1) >>> phi = (X) ** 2 + (Y) ** 2 - r ** 2 >>> speed = np.ones_like(phi) >>> speed[X>0.25] = 3.0 >>> d2, f_ext = extension_velocities(phi, speed, dx)
>>> assert (f_ext <= 3.0000001).all() >>> assert (f_ext >= 1).all()
>>> np.testing.assert_almost_equal(f_ext[137, 95], 1, 3) >>> np.testing.assert_almost_equal(f_ext[103, 78], 1, 2) >>> np.testing.assert_almost_equal(f_ext[72, 100], 3, 3) >>> np.testing.assert_almost_equal(f_ext[72, 86], 3, 3) >>> np.testing.assert_almost_equal(f_ext[110, 121], 3, 3)
Simple two point tests
>>> np.testing.assert_array_equal(distance([-1, 1]), ... [-0.5, 0.5]) >>> np.testing.assert_allclose(distance([-1, -1, -1, 1, 1, 1]), ... [-2.5, -1.5, -0.5, 0.5, 1.5, 2.5]) >>> np.testing.assert_allclose(distance([1, 1, 1, -1, -1, -1]), ... [2.5, 1.5, 0.5, -0.5, -1.5, -2.5])
Three point test case
>>> np.testing.assert_array_equal(distance([-1, 0, 1]), [-1, 0, 1]) >>> np.testing.assert_array_equal(distance([-1, 0, 1], dx=[2]), [-2, 0, 2]) >>> np.testing.assert_array_equal(distance([-1, 0, 1], dx=2), [-2, 0, 2]) >>> np.testing.assert_array_equal(distance([-1, 0, 1], dx=2.0), [-2, 0, 2]) >>> np.testing.assert_array_equal(travel_time([1, 0, -1], [1, 1, 1]), ... [1, 0, 1]) >>> np.testing.assert_array_equal(travel_time([-1, 0, 1], [1, 1, 1]), ... [1, 0, 1]) >>> np.testing.assert_array_equal(travel_time([1, 0, -1], [1, 1, 1], dx=2), ... [2, 0, 2]) >>> np.testing.assert_array_equal(travel_time([1, 0, -1], [1, 1, 1], dx=[2]), ... [2, 0, 2]) >>> np.testing.assert_array_equal(travel_time([1, 0, -1], [1, 1, 1], dx=2.0), ... [2, 0, 2])
Travel time tests 1
>>> np.testing.assert_allclose(travel_time([0, 1, 1, 1, 1], [2, 2, 2, 2, 2]), ... [0, 0.5, 1.0, 1.5, 2.0]) >>> np.testing.assert_array_equal(travel_time([1, 0, -1], [2, 2, 2]), ... [0.5, 0, 0.5])
Travel time tests 2
>>> phi = [1, 1, 1, -1, -1, -1] >>> t = travel_time(phi, np.ones_like(phi)) >>> exact = [2.5, 1.5, 0.5, 0.5, 1.5, 2.5] >>> np.testing.assert_allclose(t, exact)
Travel time tests 3
>>> phi = [-1, -1, -1, 1, 1, 1] >>> t = travel_time(phi, np.ones_like(phi)) >>> exact = [2.5, 1.5, 0.5, 0.5, 1.5, 2.5] >>> np.testing.assert_allclose(t, exact)
Corner case
>>> np.testing.assert_array_equal(distance([0, 0]), [0, 0]) >>> np.testing.assert_array_equal(travel_time([0, 0], [1, 1]), [0, 0])
Test zero
>>> distance([1, 0, 1, 1], 0) Traceback (most recent call last): ... ValueError: dx must be greater than zero
Test dx shape
>>> distance([0, 0, 1, 0, 0], [0, 0, 1, 0, 0]) Traceback (most recent call last): ... ValueError: dx must be of length len(phi.shape)
Test for small speeds
Test catching speeds which are too small. Speeds less than the machine epsilon are masked off to avoid an overflow.
>>> t = travel_time([-1, -1, 0, 1, 1], [1, 1, 1, 1, 0]) >>> assert isinstance(t, np.ma.MaskedArray) >>> np.testing.assert_array_equal(t.data[:-1], [2, 1, 0, 1]) >>> np.testing.assert_array_equal(t.mask, [False, False, False, False, True])
>>> t2 = travel_time([-1, -1, 0, 1, 1], [1, 1, 1, 1, 1e-300]) >>> np.testing.assert_array_equal(t, t2)
Mask test
Test that when the mask cuts off the solution, the cut off points are also masked.
>>> ma = np.ma.MaskedArray([1, 1, 1, 0], [False, True, False, False]) >>> d = distance(ma) >>> exact = np.ma.MaskedArray([0, 0, 1, 0], [True, True, False, False]) >>> np.testing.assert_array_equal(d.mask, exact.mask) >>> np.testing.assert_array_equal(d, exact)
Circular level set
>>> N = 50 >>> X, Y = np.meshgrid(np.linspace(-1, 1, N), np.linspace(-1, 1, N)) >>> r = 0.5 >>> dx = 2.0 / (N - 1) >>> phi = (X) ** 2 + (Y) ** 2 - r ** 2 >>> d = distance(phi, dx, self_test=True) >>> exact = np.sqrt(X ** 2 + Y ** 2) - r >>> np.testing.assert_allclose(d, exact, atol=dx)
Planar level set
>>> N = 50 >>> X, Y = np.meshgrid(np.linspace(-1, 1, N), np.linspace(-1, 1, N)) >>> dx = 2.0 / (N - 1) >>> phi = np.ones_like(X) >>> phi[0, :] = -1 >>> d = distance(phi, dx, self_test=True) >>> exact = Y + 1 - dx / 2.0 >>> np.testing.assert_allclose(d, exact)
Masked input
>>> N = 50 >>> X, Y = np.meshgrid(np.linspace(-1, 1, N), np.linspace(-1, 1, N)) >>> dx = 2.0 / (N - 1) >>> phi = np.ones_like(X) >>> phi[0, 0] = -1 >>> mask = np.logical_and(abs(X) < 0.25, abs(Y) < 0.25) >>> mphi = np.ma.MaskedArray(phi.copy(), mask) >>> d0 = distance(phi, dx, self_test=True) >>> d = distance(mphi, dx, self_test=True) >>> d0[mask] = 0 >>> d[mask] = 0 >>> shadow = d0 - d >>> bsh = abs(shadow) > 0.001 >>> diff = (bsh).sum()
>>> assert diff > 635 and diff < 645
Test Eikonal solution
>>> N = 50 >>> X, Y = np.meshgrid(np.linspace(-1, 1, N), np.linspace(-1, 1, N)) >>> r = 0.5 >>> dx = 2.0 / (N - 1) >>> phi = (X) ** 2 + (Y) ** 2 - r ** 2 >>> speed = np.ones_like(phi) * 2 >>> t = travel_time(phi, speed, dx) >>> exact = 0.5 * np.abs(np.sqrt(X ** 2 + Y ** 2) - 0.5)
>>> np.testing.assert_allclose(t, exact, atol=dx)
Test 1d
>>> N = 100 >>> X = np.linspace(-1.0, 1.0, N) >>> dx = 2.0 / (N - 1) >>> phi = np.zeros_like(X) >>> phi[X < 0] = -1 >>> phi[X > 0] = 1 >>> d = distance(phi, dx, self_test=True)
>>> np.testing.assert_allclose(d, X)
Test 3d
>>> N = 15 >>> X = np.linspace(-1, 1, N) >>> Y = np.linspace(-1, 1, N) >>> Z = np.linspace(-1, 1, N) >>> phi = np.ones((N, N, N)) >>> phi[0, 0, 0] = -1.0 >>> dx = 2.0 / (N - 1) >>> d = distance(phi, dx, self_test=True)
>>> exact = np.sqrt((X + 1) ** 2 + ... (Y + 1)[:, np.newaxis] ** 2 + ... (Z + 1)[:, np.newaxis, np.newaxis] ** 2)
>>> np.testing.assert_allclose(d, exact, atol=dx)
Test default dx
>>> N = 50 >>> X, Y = np.meshgrid(np.linspace(-1, 1, N), np.linspace(-1, 1, N)) >>> r = 0.5 >>> phi = (X) ** 2 + (Y) ** 2 - r ** 2 >>> speed = np.ones_like(phi) * 2 >>> out = travel_time(phi, speed, self_test=True)
Test non-square grid and dx different in different directions
>>> N = 50 >>> NX, NY = N, 5 * N >>> X, Y = np.meshgrid(np.linspace(-1, 1, NY), np.linspace(-1, 1, NX)) >>> r = 0.5 >>> phi = X ** 2 + Y ** 2 - r ** 2 >>> dx = [2.0 / (NX - 1), 2.0 / (NY - 1)] >>> d = distance(phi, dx, self_test=True) >>> exact = np.sqrt(X ** 2 + Y ** 2) - r
>>> np.testing.assert_allclose(d, exact, atol=1.3*max(dx))
No zero level set test
>>> distance([1, 1], self_test=False) Traceback (most recent call last): ... ValueError: the array phi contains no zero contour (no zero level set)
Shape mismatch test
>>> travel_time([-1, 1], [2]) Traceback (most recent call last): ... ValueError: phi and speed must have the same shape
Speed wrong type test
>>> travel_time([0, 0, 1, 1], 2) Traceback (most recent call last): ... ValueError: speed must be a 1D to 12-D array of doubles
dx mismatch test
>>> travel_time([-1, 1], [2, 2], [2, 2, 2, 2]) Traceback (most recent call last): ... ValueError: dx must be of length len(phi.shape)
Test c error handling
>>> distance([-1, 1], self_test=44) Traceback (most recent call last): ... ValueError: self_test must be 0 or 1
Check array type test
>>> distance(np.array(["a", "b"])) Traceback (most recent call last): ... ValueError: phi must be a 1 to 12-D array of doubles
>>> from skfmm import heap >>> h = heap(10,True) >>> h.push(0,0.2) 0 >>> h.push(1,0.3) 1 >>> h.push(2,0.1) 2 >>> h.update(1, 0.01) >>> h.pop() (1, 0.01) >>> h.pop() (2, 0.1) >>> h.pop() (0, 0.2) >>> h.empty() True >>> h.pop() Traceback (most recent call last): ... RuntimeError: heap pop error: empty heap
Test narrow optional argument.
>>> phi = np.array([-1,-1,-1,1,1,1]) >>> d = distance(phi, narrow=1.0) >>> d masked_array(data = [-- -- -0.5 0.5 -- --], mask = [ True True False False True True], fill_value = 1e+20) >>> N = 50 >>> X, Y = np.meshgrid(np.linspace(-1, 1, N), np.linspace(-1, 1, N)) >>> r = 0.5 >>> dx = 2.0 / (N - 1) >>> phi = (X) ** 2 + (Y) ** 2 - r ** 2 >>> exact = np.sqrt(X ** 2 + Y ** 2) - r
>>> d = distance(phi, dx) >>> bandwidth=5*dx >>> d2 = distance(phi, dx, narrow=bandwidth) >>> np.testing.assert_allclose(d, exact, atol=dx) >>> # make sure we get a normal array if there are no points outside the narrow band >>> np.testing.assert_allclose(distance(phi, dx, narrow=1000*dx), exact, atol=dx)
>>> assert (d2<bandwidth).all() >>> assert type(d2) is np.ma.masked_array
>>> speed = np.ones_like(phi) >>> speed[X>0] = 1.8 >>> t = travel_time(phi, speed, dx) >>> t2 = travel_time(phi, speed, dx, narrow=bandwidth) >>> assert type(t2) is np.ma.masked_array >>> assert (t2<bandwidth).all()
>>> speed = np.ones_like(phi) >>> speed = X + Y >>> ed, ev = extension_velocities(phi, speed, dx) >>> ed2, ev2 = extension_velocities(phi, speed, dx, narrow=bandwidth) >>> assert type(ed2) is np.ma.masked_array >>> assert type(ev2) is np.ma.masked_array >>> assert (ed2<bandwidth).all() >>> assert ed2.shape == ed.shape == ev.shape == ev2.shape >>> distance(phi, dx, narrow=-1) Traceback (most recent call last): ... ValueError: parameter "narrow" must be greater than or equal to zero. >>> # make sure the narrow options works with an existing mask. >>> mask = abs(X)<0.25 >>> d3 = distance(np.ma.masked_array(phi, mask), dx, narrow=bandwidth) >>> assert (d3.mask[mask]==True).all() >>> assert d3.mask.sum() > mask.sum()
Testing periodic argument
>>> X, Y = np.meshgrid(np.linspace(-2,2,200), np.linspace(-2,2,200)) >>> phi = -1*np.ones_like(X); phi[X**2+(Y-0.9)**2<0.5] = 1.0 >>> speed = np.ones_like(X); speed[(X-0.9)**2+Y**2<1.0] = 2.0 >>> np.allclose(distance(phi),distance(phi,periodic=False)) and np.allclose(distance(phi),distance(phi,periodic=(0,0))) True >>> np.allclose(travel_time(phi,speed),travel_time(phi,speed,periodic=False)) and np.allclose(travel_time(phi,speed),travel_time(phi,speed,periodic=(0,0))) True >>> phi = -1*np.ones_like(X); phi[X**2+Y**2<0.5] = 1.0 >>> speed = np.ones_like(X); speed[X**2+Y**2<1.0] = 2.0 >>> np.allclose(distance(phi),distance(phi,periodic=True)) and np.allclose(distance(phi),distance(phi,periodic=(1,1))) True >>> np.allclose(travel_time(phi,speed),travel_time(phi,speed,periodic=True)) and np.allclose(travel_time(phi,speed),travel_time(phi,speed,periodic=(1,1))) True >>> np.allclose(travel_time(phi,speed),travel_time(phi,speed,periodic=True)) and np.allclose(travel_time(phi,speed),travel_time(phi,speed,periodic=[1,1])) True >>> np.allclose(travel_time(phi,speed),travel_time(phi,speed,periodic=True)) and np.allclose(travel_time(phi,speed),travel_time(phi,speed,periodic=[True,True])) True >>> phi = -1*np.ones_like(X); phi[X**2+(Y-0.9)**2<0.5] = 1.0 >>> speed = np.ones_like(X); speed[(X-0.9)**2+Y**2<1.0] = 2.0 >>> np.allclose(distance(np.roll(phi,137,axis=0),periodic=True),np.roll(distance(phi,periodic=True),137,axis=0)) True >>> np.allclose(travel_time(np.roll(phi,-77,axis=1),np.roll(speed,-77,axis=1),periodic=True),np.roll(travel_time(phi,speed,periodic=True),-77,axis=1)) True
>>> phi=[1,-1,1,1,1,1] >>> speed=[4,1,2,2,2,2] >>> np.allclose(extension_velocities(phi,speed)[1],(2.5,2.5,1.5,1.5,1.5,1.5)) True >>> np.allclose(extension_velocities(phi,speed,periodic=True)[1],(2.5,2.5,1.5,1.5,1.5,2.5)) True