|
EcohydroLib
1.29
|
|
EcohydroLib
1.29
|
This software is provided free of charge under the New BSD License. Please see the following license information:
Copyright (c) 2013-2015 University of North Carolina at Chapel Hill All rights reserved.
Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE UNIVERSITY OF NORTH CAROLINA AT CHAPEL HILL BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
Brian Miles brian_miles@unc.edu
Lawrence E. Band lband@email.unc.edu
For questions or support contact Brian Miles
This work was supported by the following NSF grants
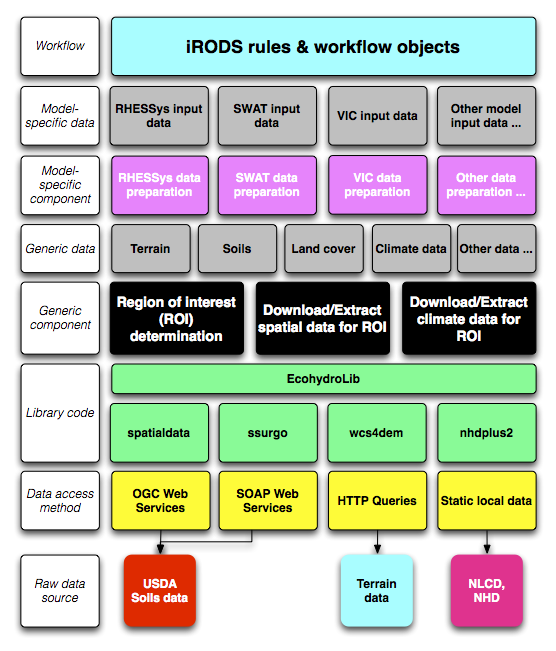
EcohydroLib provides a series of Python scripts for performing ecohydrology data preparation workflows. Workflow sub-components are orchestrated via a metadata persistence store provided by the ecohydrolib.metadata package. These scripts are built on top of a series of task-oriented APIs defined in the Python package EcohydroLib. The workflow scripts provide tools for downloading and manipulating geospatial data needed to run a ecohydrology models, information such as: digital elevation model (DEM), soils, land cover, and vegetation leaf area index. These data can be drawn both from national spatial data infrastructure (NLCD, SSURGO) as well as custom local datasets.
A metadata store is used to orchestrate a series of workflow scripts used to prepare data for an ecohydrology model. The current implementation uses the Python ConfigParser key-value storage mechanism to persist metadata to disk, however conceivably any key-value store could be used. The metadata contains information related to the study area (e.g. bounding box, spatial reference, DEM resolution), as well as provenance information for each spatial data layer imported, and a processing history that records the parameters used to invoke each workflow script; provenance information is represented as a subset of Dublin Core attributes (http://dublincore.org). When using the workflow scripts in a stand-alone environment, the metadata store provides information necessary to understand where ecohydrology input data came from, and what transformations have been made to the data. When the workflow scripts are integrated into a data grid or workflow environment (e.g. iRODS), the metadata store can serve as a staging area for metadata and provenance information that will be registered into the formal workflow environment.
The fundamental operation for any ecohydrology modeling workflow is to define the study region of interest (ROI). In EcohydroLib the ROI is simply defined as a bounding box of WGS84 latitude and longitude coordinates (e.g. coordinates for the upper- left and lower-right corners). For workflows using the National Hydrography Dataset (NHD), the ROI bounding box can be derived using catchment polygons associated with the stream reaches upstream of a particular gage. The user begins by picking a streamflow discharge gage listed in the NHD dataset. EcohydroLib can then determine the stream reaches upstream of the data, and then select the catchment polygons associated with each upstream reach. From these polygons, the bounding box of the land area draining through the streamflow gage can easily be calculated.
Once the ROI is known, EcohydroLib can extract datasets (DEM, soils, etc.) specific to the study area. Some of these datasets are extracted from static local copies of national spatial data (e.g. NLCD), while other are retrieved via web services interfaces from federal agency data centers (e.g. SSURGO soils data from USDA) or from third-party data centers (GeoBrain's DEM Explorer). However it is also possible for the user to register their own custom data for a given datatype (e.g. local LIDAR-based DEM).
Source code can be found at: https://github.com/selimnairb/EcohydroLib.
Documentation can be found at: http://pythonhosted.org/ecohydrolib
Detailed installation and usage instructions can be found in the RHESSysWorkflows readme.
The following instructions should only be used by advanced users.
Using easy_install:
easy_install –script-dir /path/to/install/scripts EcohydroLib
Using pip:
pip install EcohydroLib
It is recommended that you install the workflow scripts in a location distinct from where the Python package will be installed. This is accomplished by specifying the –script-dir option to easy install (see above).
Note, pyspatialite 3.0.1, needed for GHCNDSetup.py and GetGHCNDailyClimateData*.py, currently fails to build under easy_install/pip. Until this is fixed by the pyspatialite developer, I have removed pyspatialite from the dependency list. If you need to use GNCHD data, you can install pyspatialite manually using the following steps (this can be done before or after installing EcohydroLib):
Python 2.7
Libraries (with headers):
Binaries:
Before EcohydroLib is able to extract study area ROI using the NHDPlus dataset, it is necessary to have a local copy of the NHDPlus dataset. Owing to the large size of the NHDPlus dataset, these data are distributed as as series of compressed archives broken into several regions for the continental U.S. There are two choices for obtaining NHDPlus in a format usable by EcohydroLib (as several SQLite3 databases). A national-scale dataset (i.e. covering the entire continental U.S.) is available for download here:
Once downloaded, extract the archive and record its location in your EcohydroLib configuation file; see the 'Configuration files' section for more information.
If you wish to build you own copy of the database (i.e. for a subset of U.S. country) a script for building the dataset from downloaded NHDPlus V2 7z archives is provided in bin/NHDPlusV2Setup/NHDPlusV2Setup.py. The following NHDPlus V2 datasets are required:
Note that the NHDPlusAttributes, NHDPlusCatchment, and NHDPlusSnapshot data are released as regional subsets (due to the large size and complexity of the data). NHDPlusV2Setup.py can build its NHDPlus SQLite3 databases for any number of regions; all data for the desired number of regions will be combined into a single database.
Once you've decided which NHDPlusV2 regions you wish to build a database for, simply download the relevant 7z archives from the NHDPlusV2 web site (see above), and store the archives in a single directory. NHDPlusV2Setup.py will unpack these archives into a specified output location and then will process the unarchived files into the following databases: - Catchment.sqlite (a spatial dataset containing all catchment polygons in the selected NHD region(s); - GageLoc.sqlite (a spatial dataset containing streamflow gage points for the national NHD dataset; - NHDPlusDB.sqlite (a tabular dataset containing other NHD data needed by EcohydroLib).
Make sure to edit your configuration file to include the absolute paths of these files (see below).
For national NHD coverage, Catchment.sqlite is over 8 GB, and NHDPlusDB.sqlite is over 2 GB, so you will need a kernel and filesystem that has large file support to build and use these datasets. Also, it may take over an hour to create these datasets; 8 GB of memory or more is recommended to build the datasets efficiently. However, database setup is a one-time process, and you can use databases created on one machine on other machines, provided SQLite3 is installed. NHDPlusV2Setup.py creates each database with the indices needed by EcohydroLib, so lookups are very fast.
To use HYDRO1k basin shapefile, you must first uncompress na_bas.e00.gz to na_bas.e00. Then you must convert the e00 (Arc interchange file) to a shapefile using a tool such as ArcGIS.
To download NCDC Global Historical Climatology Network (GHCN) dataset for daily climate data, you must first create the spatialite database that EcohydroLib uses to find climate stations using spatial queries. This database is created using bin/GHCNDSetup/GHCNDSetup.py. The output from the script will be a spatialite database. Make sure to edit your configuration file and set PATH_OF_STATION_DB to the absolute path of this spatialite database (see below).
Many of the command line scripts (including NHDPlusV2Setup.py) require a configuration file to specify locations to executables and datasets required by the ecohydrology workflow libraries. The configuration file can be specified via the environmental variable ECOHYDROLIB_CFG or via command line option. Here is an example configuration file:
[GDAL/OGR]
PATH_OF_OGR2OGR = /Library/Frameworks/GDAL.framework/Versions/Current/Programs/ogr2ogr
PATH_OF_GDAL_RASTERIZE = /Library/Frameworks/GDAL.framework/Versions/Current/Programs/gdal_rasterize
PATH_OF_GDAL_WARP = /Library/Frameworks/GDAL.framework/Versions/Current/Programs/gdalwarp
PATH_OF_GDAL_TRANSLATE = /Library/Frameworks/GDAL.framework/Versions/Current/Programs/gdal_translate
[NHDPLUS2]
PATH_OF_NHDPLUS2_DB = /Users/<username>/Research/data/GIS/NHDPlusV21/national/NHDPlusDB.sqlite
PATH_OF_NHDPLUS2_CATCHMENT = /Users/<username>/Research/data/GIS/NHDPlusV21/national/Catchment.sqlite
PATH_OF_NHDPLUS2_GAGELOC = /Users/<username>/Research/data/GIS/NHDPlusV21/national/GageLoc.sqlite
[SOLIM]
PATH_OF_SOLIM = /Users/<username>/Research/bin/solim/solim.out
[NLCD]
PATH_OF_NLCD2006 = /Users/<username>/Research/data/GIS/NLCD2006/nlcd2006/nlcd2006_landcover_4-20-11_se5.img
[HYDRO1k]
PATH_OF_HYDRO1K_DEM = /Users/<username>/Research/data/GIS/HYDRO1k/na/na_dem.bil
PATH_OF_HYDRO1K_BAS = /Users/<username>/Research/data/GIS/HYDRO1k/na/na_bas_polygon.shp
HYDRO1k_BAS_LAYER_NAME = na_bas_polygon
[GHCND]
PATH_OF_STATION_DB = /Users/<username>/Research/data/obs/NCDC/GHCND/GHCND.spatialite
[UTIL]
PATH_OF_FIND = /usr/bin/find
PATH_OF_SEVEN_ZIP = /opt/local/bin/7z
PATH_OF_SQLITE = /opt/local/bin/sqlite3
If you create your initial configuration file by copying and pasting from this documentation, make sure to remove any leading spaces from each line of the file.
A workflow using data from large-scale spatial data infrastructure will consist of running the follow scripts in the following order:
The first 4 steps must be run in this order, the remaining workflow components can be run in any order. Other workflow components, e.g. to register a custom dataset, can be substituted for the latter 4 workflow components as well (as indicated above). See the documentation for each script to see invocations details.
A workflow collecting data appropriate for large-scale land surface process models may consist of running the following scripts in the following order:
A workflow using custom local data sources will consist of running the follow scripts in the following order:
A workflow using custom streamflow gage, but with standard spatial data (NED, NLCD, SSURGO) could consist of running the follow scripts in the following order:
 1.8.9.1
1.8.9.1