For reading, manipulating and plotting the output files from DDSCAT
Results from an individual file (e.g. qtable or shape.dat) are held within a dictionary-like object. The columns of the table (e.g. wavelength, Q_ext) are accessible as entires in the dictionary that can be accessed using the key name (e.g. T[‘wave’], T[‘Q_ext’]). Other results parameters like polarization are available as attribute fields (eg. T.Epol). The available fields vary between result types.
The fields available as table columns can be found with T.keys(). The other attributes can be found with dir(T).
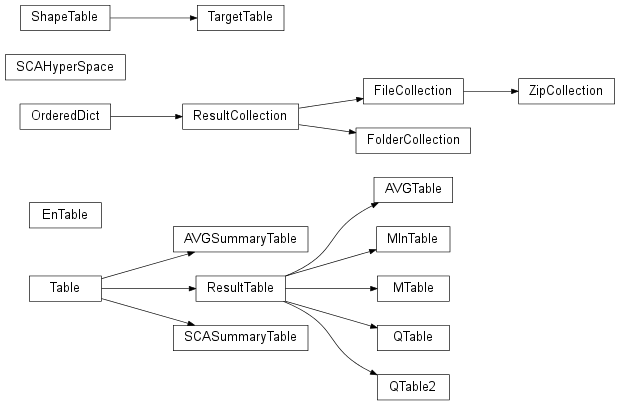
| Table() | Base class for tables |
| ResultTable(fname, hdr_len, c_width[, ...]) | Base class for results tables read from file. |
| AVGTable([fname, folder]) | A class for reading .avg files. |
| AVGSummaryTable([fname, folder, npol, ...]) | A class for reading the summary section of AVGTables. |
| SCASummaryTable([fname, folder, npol, zfile]) | A class for reading the summary section of .sca files. |
| QTable([fname, folder]) | A class for reading qtable output files. |
| QTable2([fname, folder]) | A class for reading qtable2 output files. |
| MTable([fname]) | A class for reading mtables output by DDSCAT. |
| MInTable([fname]) | A class for reading material input files. |
| ShapeTable([fname, folder]) | A class for reading shape.dat files. |
| TargetTable([fname, folder]) | A class for reading target.out files. |
| EnTable([fname, folder, zfile]) | Class for reading the .E1 and .E2 files from nearfield calcs. |
| ResultCollection() |
| ResultCollection() | |
| FolderCollection([r_type, path, recurse, fields]) | |
| FileCollection([r_type, path]) | |
| ZipCollection([r_type, folder]) | |
| SCAHyperSpace() | Create an object that stores the results from an isotropic simulation into a multi-dimensional np array. |
| _dichroism_calculator(L, R) | Calculates the difference spectrum of two spectra. |
| split_string(s[, widths]) | Splits a string into list of strings at the partition points provided by the sequence widths. |
| clean_string(s) | Remove illegal tokens from a string to return something appropriate for a Python name |
| Esq(E) | Return the magnitude squared of a vector field. |
For reading, manipulating and plotting the output files from DDSCAT
Results from an individual file (e.g. qtable or shape.dat) are held within a dictionary-like object. The columns of the table (e.g. wavelength, Q_ext) are accessible as entires in the dictionary that can be accessed using the key name (e.g. T[‘wave’], T[‘Q_ext’]). Other results parameters like polarization are available as attribute fields (eg. T.Epol). The available fields vary between result types.
The fields available as table columns can be found with T.keys(). The other attributes can be found with dir(T).
Bases: ScatPy.results.Table
A class for reading the summary section of AVGTables.
Bases: ScatPy.results.ResultTable
A class for reading .avg files.
Bases: dict
Class for reading the .E1 and .E2 files from nearfield calcs.
| Parameters: |
|
|---|
Results are returned as a dictionary of complex 3D numpy arrays. The key names corresponds to the following fields:
Comp: composition identifier at all points (0 for vacuum) Pol: polarization/d^3 at all points Esca: complex radiated E field at all points Einc: complex incident E field at all points Pdia: diagonal element of polarizability/d^3 at all pts Etot: The complex total E field at all points Etot2: The magnitude squared of the E field at all points
Bases: ScatPy.results.ResultTable
A class for reading material input files.
Simple file names are resolved relative to the default material_library path.
This class can be called to return an interpolated value at a requested wavelength. e.g.:
M=MInTable('Gold.txt')
refr_index=M(0.750)
Bases: ScatPy.results.ResultTable
A class for reading mtables output by DDSCAT.
Fields are: wave(um), f(cm-1), Re(m), Im(m), Re(eps), Im(eps)
Bases: ScatPy.results.ResultTable
A class for reading qtable output files.
Fields are: aeff, wave, Q_ext, Q_abs, Q_sca, g(1)=<cos>, <cos^2>, Q_bk, Nsca
Bases: ScatPy.results.ResultTable
A class for reading qtable2 output files.
Fields are: aeff, wave, Q_pha, Q_pol, Q_cpol
Bases: collections.OrderedDict
Calculate the dichroism between pairs of spectra.
Spectra are considered to form a pair if they come from folders with identical names suffixed with _cL and _cR (e.g. ‘sphere1_cL’ and ‘sphere2_cR’).
Bases: ScatPy.results.Table
Base class for results tables read from file.
| Parameters: |
|
|---|
Most DDscat output files have fixed column widths and cannot be relied upon to have whitespace between adjacent columns. Therefore the columns’ width must be provided in the c_width list.
Field names are derived from the column headings in the result table. The fields can be accessed like a Python dict, e.g. T[‘wave’]
Create an object that stores the results from an isotropic simulation into a multi-dimensional np array.
The array is addressed by integer indices. The wavelength/radius/angle corresponding to a given index can be recoverred with he lists w_range, r_range, etc.
Bases: ScatPy.results.Table
A class for reading the summary section of .sca files.
Bases: dict
A class for reading shape.dat files.
Bases: dict
Base class for tables
Results from an individual file (e.g. qtable or shape.dat) are held within a dictionary-like object. The columns of the table (e.g. wavelength, Q_ext) are accessible as entires in the dictionary that can be accessed using the key name (e.g. T[‘wave’], T[‘Q_ext’]). Other results parameters like polarization are available as attribute fields (eg. T.Epol). The available fields vary between result types.
The fields available as table columns can be found with T.keys(). The other attributes can be found with dir(T).
Tables have attributes `x_field` and `y_fields` which indicate the default fields to use for the x axis and y axes when plotting.
Export the table as an ascii file
| Parameters: |
|
|---|
Interpolates the data onto a new (denser) xrange.
Data is modified in place.
Plot the table contents.
| Parameters: |
|
|---|---|
| Returns: | the axes of the plot |
Bases: ScatPy.results.ShapeTable
A class for reading target.out files.
Remove illegal tokens from a string to return something appropriate for a Python name
Calculate the ellipticity in deg of a suspension
| Parameters: |
|
|---|
Calculate the ellipticity in deg of a suspension
| Parameters: |
|
|---|