Expression Profile Distances¶

Computes distances between gene expression levels.
Signals¶
Inputs:
Data
Data set.
Outputs:
Distances
Distance matrix.
Sorted Data
Data with groups as attributes.
Description¶
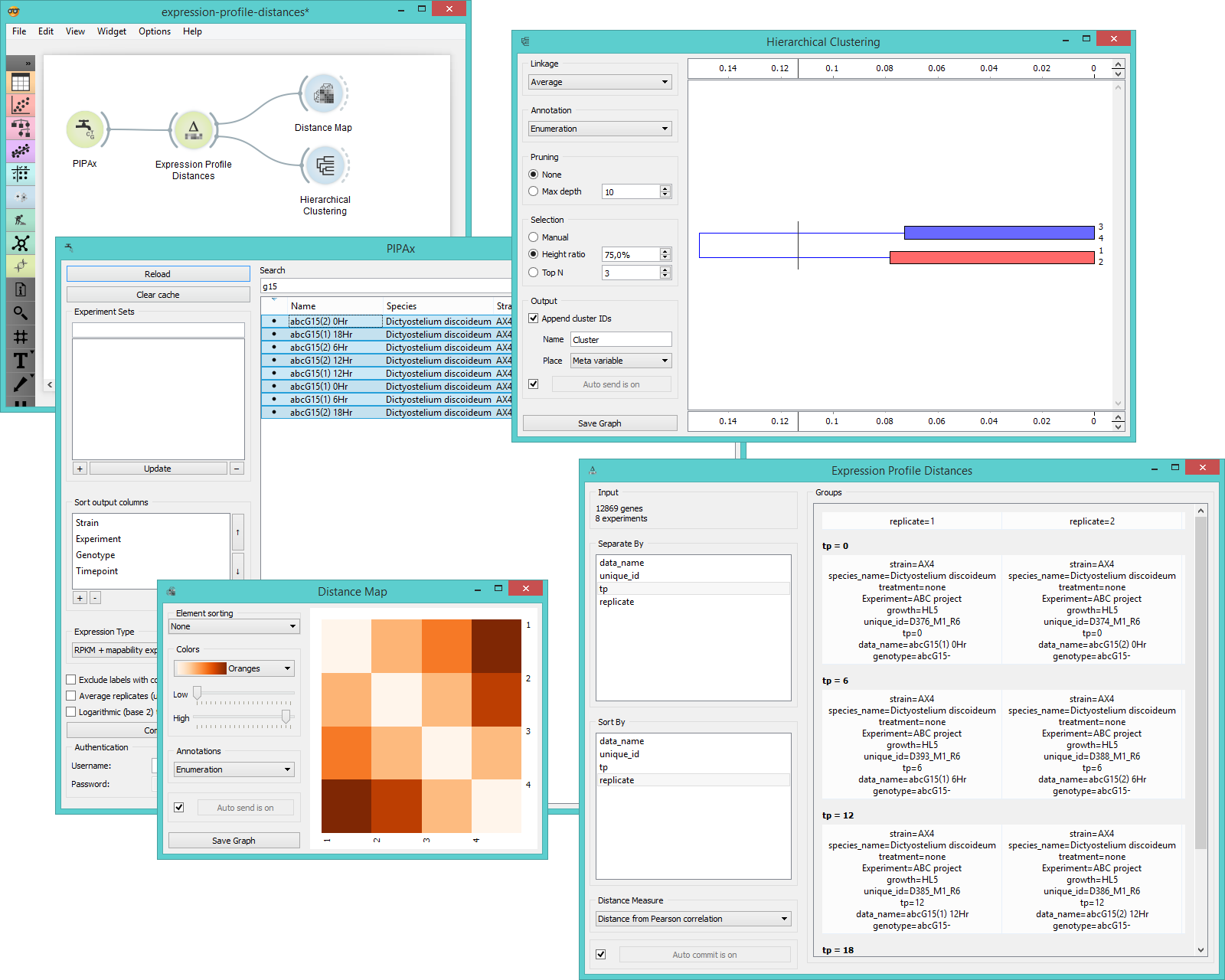
Widget Expression Profile Distances computes distances between expression levels among groups of data. Groups are data clusters set by the user through separate by function in the widget. Data can be separated by one or more variable labels (usually timepoint, replicates, IDs, etc.). Widget outputs distance matrix that can be fed into Distance Map and Hierarchical Clustering widgets.
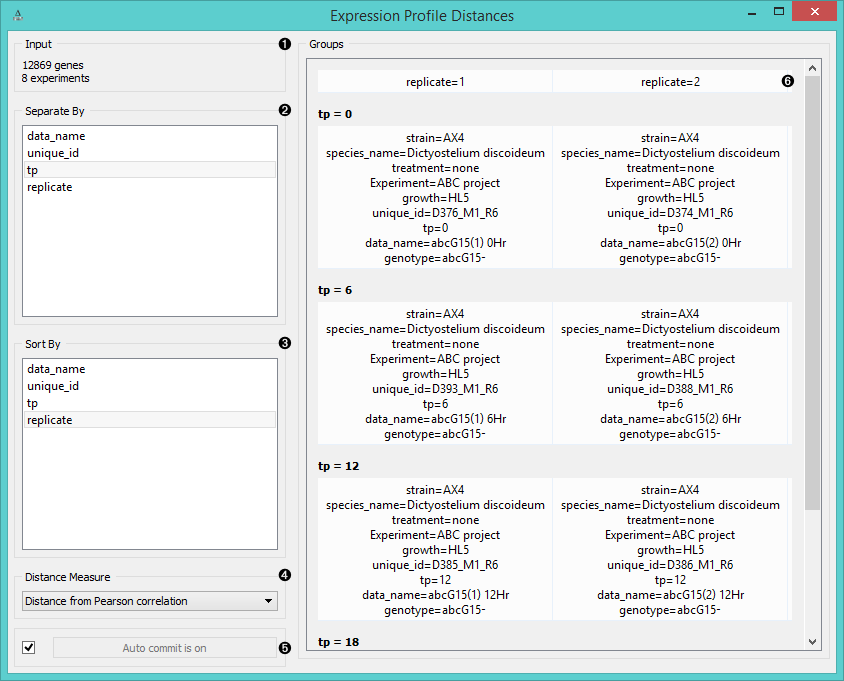
- Information on the input data.
- Separate the experiments into groups by labels (normally timepoint, replicates, data name, etc.).
- Sort the experiments inside the group by labels.
- Choose the Distance Measure:
- If Auto commit is on, the widget will automatically compute the distances and output them. Alternatively click Commit.
- This snapshot shows 4 groups of experiments (tp=0, tp=6, tp=12, tp=18) with 2 experiments (replicates) in each group.
Example¶
Expression Profile Distances widget is used to calculate distances between gene expression values sorted by labels. We chose 8 experiments measuring gene expression levels on Dictyostelium discoideum at different timepoints. In the Expression Profile Distances widget we separated the data by timepoint and sorted them by replicates. We could see the grouping immediately in the Groups box on the right. Then we fed the results to Distance Map and Hierarchical Clustering to visualize the distances and cluster the attributes.