evaluate Package¶
evaluate Package¶
evaluate¶
This package deals with the evaluation of the parametrized models returned by optimization routines. Evaluation criteria used are:
- Voltage mean square error.
- Spike distance metrics.
- Gamma coincidence factor - spkd_lib.gamma_factor()
- Victor Papura spike distance - spkd_lib.victor_purpura_dist()
- van Rossum distance - spkd_lib.van_rossum_dist()
- Schreiber et al. similarity measure - spkd_lib.schreiber_sim()
evaluate Module¶
This simulation file runs the subthreshold model together with the threshold model and plots the results against the actual recordings.
- fit_neuron.evaluate.evaluate.plot_sim_vs_real(simulated_voltage_dict=[], bio_voltage_dict=[], input_current_dict=[], plot_nan_as_spike=True, fig_dir='./', file_ext='.png')[source]¶
Plots the simulated voltage traces against the actual voltage traces and saves the result as .eps file.
Parameters: - bio_voltage_dict – dictionary, values of which are arrays of voltage traces
- input_voltage_dict – dictionary of input current injections, values of which are arrays of input stimuli
- simulated_voltage_dict – dictionary. values of which are lists of arrays of monte carlo simulated voltage traces
- fig_dir – directory into which figures will be plotted
- nan_as_spike (bool) – do we draw spiking lines where the traces are nan?
- file_ext (str) – file extension of plotting results
Note
It is assumed that the keys of the dictionary inputs correspond to each other.
- fit_neuron.evaluate.evaluate.plot_spk_performance_metrics(bio_voltage_dict, simulated_voltage_dict, fig_dir='./', file_ext='.png', dt=0.0001)[source]¶
Plots spike distance metrics for the data provided. Metrics used are the gamma coincidence factor, the Schreiber similarity, and the van Rossum distance. Each is plotted as a function of the smoothness parameter for that particular metric. The spike times are extracted from the voltage traces using fit_neuron.data.extract_spikes.spk_from_sim() and fit_neuron.data.extract_spikes.spk_from_bio().
Parameters: - bio_voltage_dict (dict) – recorded voltage traces, indexed by a file ID
- simulated_voltage_dict (dict) – simulated voltage traces, indexed by a file ID
- fig_dir – directory where the plots will be saved
- file_ext – file extension
- dt – time step interval
Note
The keys of bio_voltage_dict and simulated_voltage_dict must match. In addition, there may be multiple simulations for a single file ID, but there should be a single recorded voltage trace for a file ID.
- fit_neuron.evaluate.evaluate.simulate(neuron=None, input_current_list=[], membrane_voltage_list=[], file_id_list=[], reps=1)[source]¶
[simulated_voltage_list,sim_file_id_list] = evaluate.simulate(neuron,input_current_list,file_id_list)
This function takes a neuron fitted from some data and simulates it against the data it is supposed to fit.
Parameters: - neuron – a neuron instance
- input_current_list – list of input currents
- membrane_voltage_list – list of voltage trace, useful for initializing neuron
- file_id_list – list of file ID’s corresponding to input_current_list
- reps – number of times we want to produce simulated voltage traces for each input current in input current list
Returns: dictionary of {file_id: simulated_voltage_list}
Note
The values in the dictionary will be lists that have lengths corresponding to reps. Each element in these lists will be an array, computed as a single monte carlo trial.
spkd_lib Module¶
spkd_lib¶
This module is a library of spike distance metrics. For more information about the spike distance metrics used below, see http://www.scholarpedia.org/article/Measures_of_spike_train_synchrony.
- class fit_neuron.evaluate.spkd_lib.ExpDecay(k=None, dt=0.0001)[source]¶
Exponentially decaying function with additive method. Useful for efficiently computing Van Rossum distance.
- fit_neuron.evaluate.spkd_lib.bivariate_spike_distance(t1, t2, ti, te, N)[source]¶
TODO: test this function
This Python code (including all further comments) was written by Jeremy Fix (see http://jeremy.fix.free.fr/), based on Matlab code written by Thomas Kreuz.
The SPIKE-distance is described in this paper:
[KT2013] Kreuz T, Chicharro D, Houghton C, Andrzejak RG, Mormann F: Monitoring spike train synchrony. J Neurophysiol 109, 1457-1472 (2013). Computes the bivariate SPIKE distance of Kreuz et al. (2012)
Parameters: - t1 – 1D array with the spiking times of two neurons.
- t2 – 1D array with the spiking times of two neurons.
- ti – beginning of time interval.
- te – end of time intervals.
- N – number of samples.
Returns: Array of the values of the distance between time ti and te with N samples.
Note
The arrays t1, t2 and values ti, te are unit less
- fit_neuron.evaluate.spkd_lib.find_corner_spikes(t, train, ibegin, ti, te)[source]¶
Return the times (t1,t2) of the spikes in train[ibegin:] such that t1 < t and t2 >= t.
- fit_neuron.evaluate.spkd_lib.gamma_factor(source, target, delta, normalize=True, dt=None)[source]¶
Returns the gamma precision factor between source and target trains, with precision delta. See [RJ2008] for a more detailed description. If normalize is True, the function returns the normalized gamma factor (less than 1.0), otherwise it returns the number of coincidences. dt is the precision of the trains, by default it is defaultclock.dt
Parameters: - source – array of spike times
- target – array of spike times
- delta – precision of coincidence detector
[RJ2008] Jolivet, Renaud, et al. “A benchmark test for a quantitative assessment of simple neuron models.” Journal of neuroscience methods 169.2 (2008): 417-424.
- fit_neuron.evaluate.spkd_lib.get_gamma_factor(coincidence_count, model_length, target_length, target_rates, delta)[source]¶
- fit_neuron.evaluate.spkd_lib.multivariate_spike_distance(spike_trains, ti, te, N)[source]¶
t is an array of spike time arrays ti the initial time of the recordings te the end time of the recordings N the number of samples used to compute the distance spike_trains is a list of arrays of shape (N, T) with N spike trains The multivariate distance is the instantaneous average over all the pairwise distances
- fit_neuron.evaluate.spkd_lib.schreiber_sim(st_0, st_1, bin_width=0.0001, sigma=0.1, t_extra=0.5)[source]¶
Computes Schreiber similarity between two spike trains as described in [SS2003].
Parameters: - st_0 – array of spike times in seconds
- st_1 – second array of spike times in seconds
- bin_width – precision in seconds over which Gaussian convolution is computed
- sigma – bandwidth of Gaussian kernel
- t_extra – how much more time in seconds after last signal do we keep convolving?
[SS2003] Schreiber, S., et al. “A new correlation-based measure of spike timing reliability.” Neurocomputing 52 (2003): 925-931.
- fit_neuron.evaluate.spkd_lib.van_rossum_dist(st_0, st_1, tc=1000, bin_width=0.0001, t_extra=1)[source]¶
Calculates the Van Rossum distance between spike trains as defined in [VR2001]. Note that the default parameters are optimized for inputs in units of seconds.
Parameters: - st_0 – array of spike times for first spike train
- st_1 – array of spike times for second spike train
- bin_width – precision in units of time to compute integral
- t_extra – how much beyond max time do we keep integrating until? This is necessary because the integral cannot in practice be evaluated between
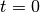 and
and 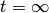 .
.
[VR2001] van Rossum, Mark CW. “A novel spike distance.” Neural Computation 13.4 (2001): 751-763.
- fit_neuron.evaluate.spkd_lib.victor_purpura_dist(tli, tlj, cost=1)[source]¶
d=spkd(tli,tlj,cost) calculates the “spike time” distance as defined [DA2003] for a single free parameter, the cost per unit of time to move a spike.
Parameters: - tli – vector of spike times for first spike train
- tlj – vector of spike times for second spike train
- cost – cost per unit time to move a spike
Returns: spike distance metric
Translated to Python by Nicolas Jimenez from Matlab code by Daniel Reich.
[DA2003] Aronov, Dmitriy. “Fast algorithm for the metric-space analysis of simultaneous responses of multiple single neurons.” Journal of Neuroscience Methods 124.2 (2003): 175-179. Here, the distance is 1 because there is one extra spike to be deleted at the end of the the first spike train:
>>> spike_time([1,2,3,4],[1,2,3],cost=1) 1
Here the distance is 1 because we shift the first spike by 0.2, leave the second alone, and shift the third one by 0.2, adding up to 0.4:
>>> spike_time([1.2,2,3.2],[1,2,3],cost=1) 0.4
Here the third spike is adjusted by 0.5, but since the cost per unit time is 0.5, the distances comes out to 0.25:
>>> spike_time([1,2,3,4],[1,2,3,3.5],cost=0.5) 0.25