Model Selection¶
Comparing machine learning models with Scikit-Learn and Yellowbrick¶
In this tutorial, we are going to look at scores for a variety of Scikit-Learn models and compare them using visual diagnostic tools from Yellowbrick in order to select the best model for our data.
About the Data¶
This tutorial uses a modified version of the mushroom dataset from the UCI Machine Learning Repository. Our objective is to predict if a mushroom is poisonous or edible based on its characteristics.
The data include descriptions of hypothetical samples corresponding to 23 species of gilled mushrooms in the Agaricus and Lepiota Family. Each species was identified as definitely edible, definitely poisonous, or of unknown edibility and not recommended (this latter class was combined with the poisonous one).
Our file, “agaricus-lepiota.txt,” contains information for 3 nominally valued attributes and a target value from 8124 instances of mushrooms (4208 edible, 3916 poisonous).
Let’s load the data with Pandas.
import os
import pandas as pd
names = [
'class',
'cap-shape',
'cap-surface',
'cap-color'
]
mushrooms = os.path.join('data','agaricus-lepiota.txt')
dataset = pd.read_csv(mushrooms)
dataset.columns = names
dataset.head()
| class | cap-shape | cap-surface | cap-color | |
|---|---|---|---|---|
| 0 | edible | bell | smooth | white |
| 1 | poisonous | convex | scaly | white |
| 2 | edible | convex | smooth | gray |
| 3 | edible | convex | scaly | yellow |
| 4 | edible | bell | smooth | white |
features = ['cap-shape', 'cap-surface', 'cap-color']
target = ['class']
X = dataset[features]
y = dataset[target]
Feature Extraction¶
Our data, including the target, is categorical. We will need to change these values to numeric ones for machine learning. In order to extract this from the dataset, we’ll have to use Scikit-Learn transformers to transform our input dataset into something that can be fit to a model. Luckily, Sckit-Learn does provide a transformer for converting categorical labels into numeric integers: sklearn.preprocessing.LabelEncoder. Unfortunately it can only transform a single vector at a time, so we’ll have to adapt it in order to apply it to multiple columns.
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
class EncodeCategorical(BaseEstimator, TransformerMixin):
"""
Encodes a specified list of columns or all columns if None.
"""
def __init__(self, columns=None):
self.columns = [col for col in columns]
self.encoders = None
def fit(self, data, target=None):
"""
Expects a data frame with named columns to encode.
"""
# Encode all columns if columns is None
if self.columns is None:
self.columns = data.columns
# Fit a label encoder for each column in the data frame
self.encoders = {
column: LabelEncoder().fit(data[column])
for column in self.columns
}
return self
def transform(self, data):
"""
Uses the encoders to transform a data frame.
"""
output = data.copy()
for column, encoder in self.encoders.items():
output[column] = encoder.transform(data[column])
return output
Modeling and Evaluation¶
Common metrics for evaluating classifiers¶
Precision is the number of correct positive results divided by the number of all positive results (e.g. How many of the mushrooms we predicted would be edible actually were?).
Recall is the number of correct positive results divided by the number of positive results that should have been returned (e.g. How many of the mushrooms that were poisonous did we accurately predict were poisonous?).
The F1 score is a measure of a test’s accuracy. It considers both the precision and the recall of the test to compute the score. The F1 score can be interpreted as a weighted average of the precision and recall, where an F1 score reaches its best value at 1 and worst at 0.
precision = true positives / (true positives + false positives)
recall = true positives / (false negatives + true positives)
F1 score = 2 * ((precision * recall) / (precision + recall))
Now we’re ready to make some predictions!
Let’s build a way to evaluate multiple estimators – first using traditional numeric scores (which we’ll later compare to some visual diagnostics from the Yellowbrick library).
from sklearn.metrics import f1_score
from sklearn.pipeline import Pipeline
def model_selection(X, y, estimator):
"""
Test various estimators.
"""
y = LabelEncoder().fit_transform(y.values.ravel())
model = Pipeline([
('label_encoding', EncodeCategorical(X.keys())),
('one_hot_encoder', OneHotEncoder()),
('estimator', estimator)
])
# Instantiate the classification model and visualizer
model.fit(X, y)
expected = y
predicted = model.predict(X)
# Compute and return the F1 score (the harmonic mean of precision and recall)
return (f1_score(expected, predicted))
# Try them all!
from sklearn.svm import LinearSVC, NuSVC, SVC
from sklearn.neighbors import KNeighborsClassifier
from sklearn.linear_model import LogisticRegressionCV, LogisticRegression, SGDClassifier
from sklearn.ensemble import BaggingClassifier, ExtraTreesClassifier, RandomForestClassifier
model_selection(X, y, LinearSVC())
0.65846308387744845
model_selection(X, y, NuSVC())
0.63838842388991346
model_selection(X, y, SVC())
0.66251459711950167
model_selection(X, y, SGDClassifier())
0.69944182052382997
model_selection(X, y, KNeighborsClassifier())
0.65802139037433149
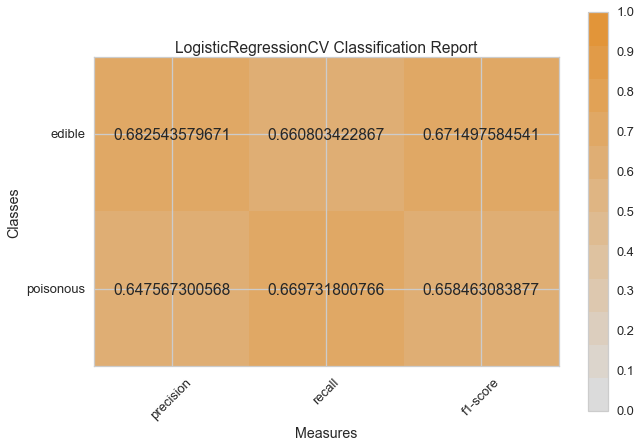
model_selection(X, y, LogisticRegressionCV())
0.65846308387744845
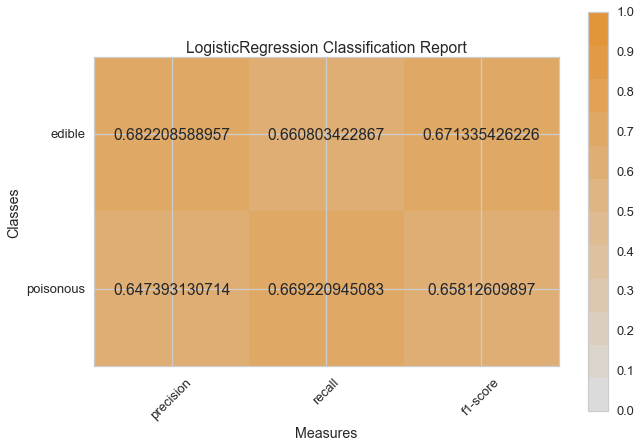
model_selection(X, y, LogisticRegression())
0.65812609897010799
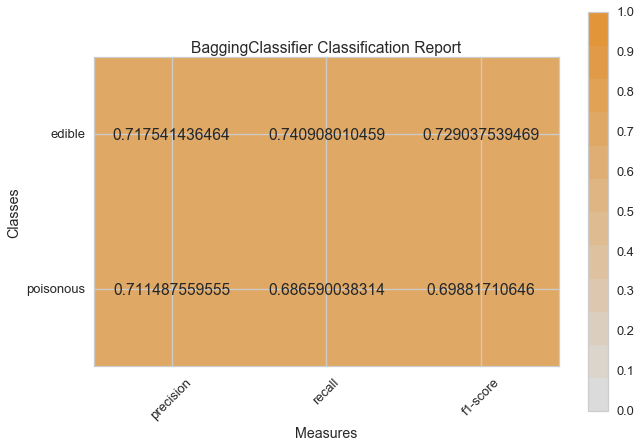
model_selection(X, y, BaggingClassifier())
0.687643484132343
model_selection(X, y, ExtraTreesClassifier())
0.68713648045448383
model_selection(X, y, RandomForestClassifier())
0.69317131158367451
Preliminary Model Evaluation¶
Based on the results from the F1 scores above, which model is performing the best?
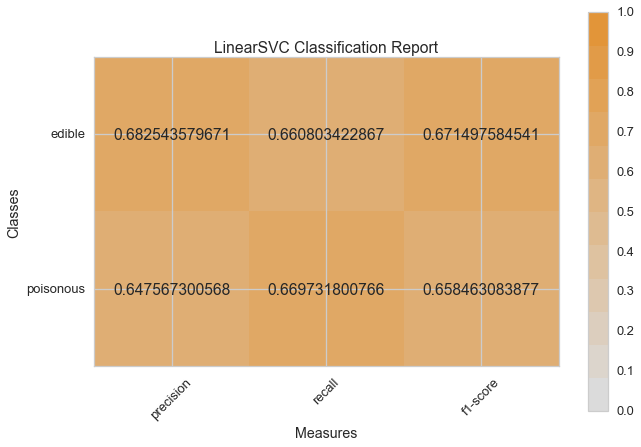
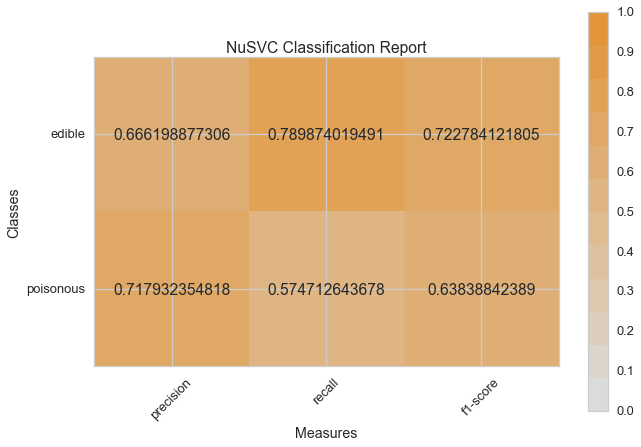
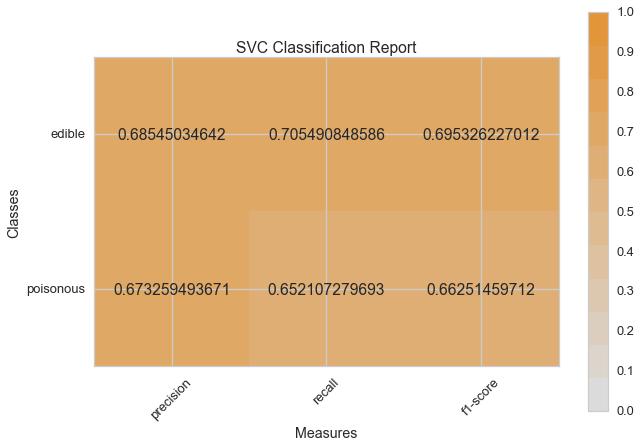
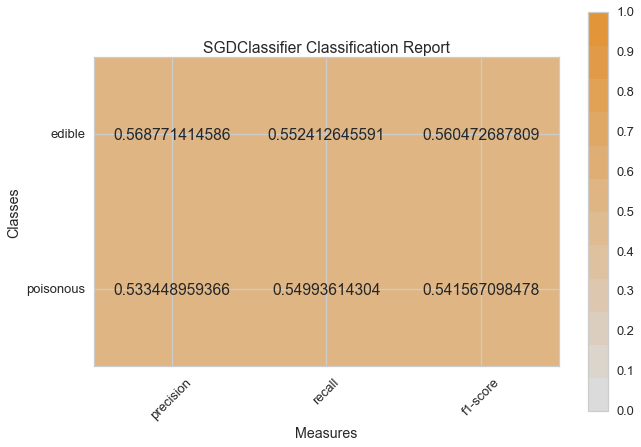
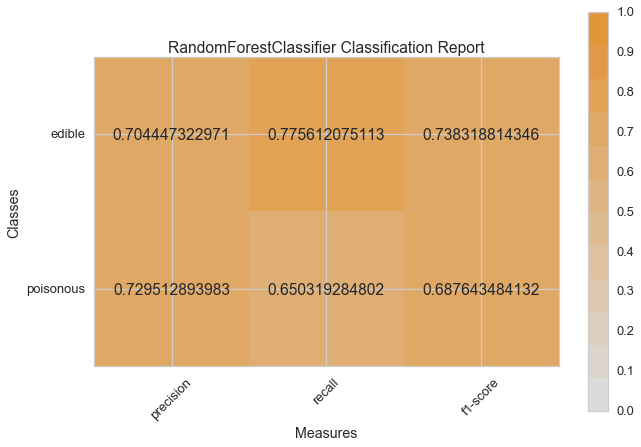
Visual Model Evaluation¶
Now let’s refactor our model evaluation function to use Yellowbrick’s
ClassificationReport class, a model visualizer that displays the
precision, recall, and F1 scores. This visual model analysis tool
integrates numerical scores as well color-coded heatmap in order to
support easy interpretation and detection, particularly the nuances of
Type I and Type II error, which are very relevant (lifesaving, even) to
our use case!
Type I error (or a “false positive”) is detecting an effect that is not present (e.g. determining a mushroom is poisonous when it is in fact edible).
Type II error (or a “false negative”) is failing to detect an effect that is present (e.g. believing a mushroom is edible when it is in fact poisonous).
from sklearn.pipeline import Pipeline
from yellowbrick.classifier import ClassificationReport
def visual_model_selection(X, y, estimator):
"""
Test various estimators.
"""
y = LabelEncoder().fit_transform(y.values.ravel())
model = Pipeline([
('label_encoding', EncodeCategorical(X.keys())),
('one_hot_encoder', OneHotEncoder()),
('estimator', estimator)
])
# Instantiate the classification model and visualizer
visualizer = ClassificationReport(model, classes=['edible', 'poisonous'])
visualizer.fit(X, y)
visualizer.score(X, y)
visualizer.poof()
visual_model_selection(X, y, LinearSVC())
visual_model_selection(X, y, NuSVC())
visual_model_selection(X, y, SVC())
visual_model_selection(X, y, SGDClassifier())
visual_model_selection(X, y, KNeighborsClassifier())
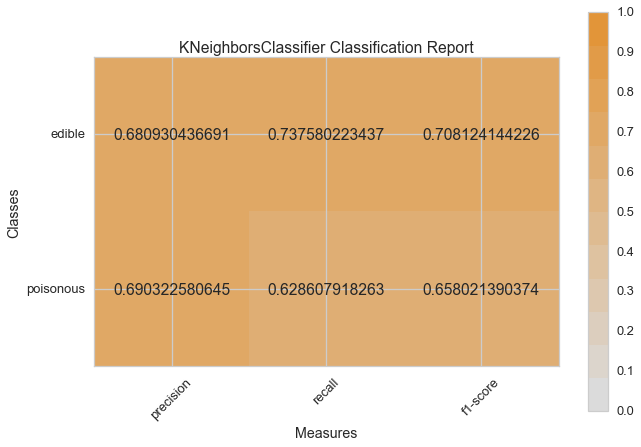
visual_model_selection(X, y, LogisticRegressionCV())
visual_model_selection(X, y, LogisticRegression())
visual_model_selection(X, y, BaggingClassifier())
visual_model_selection(X, y, ExtraTreesClassifier())
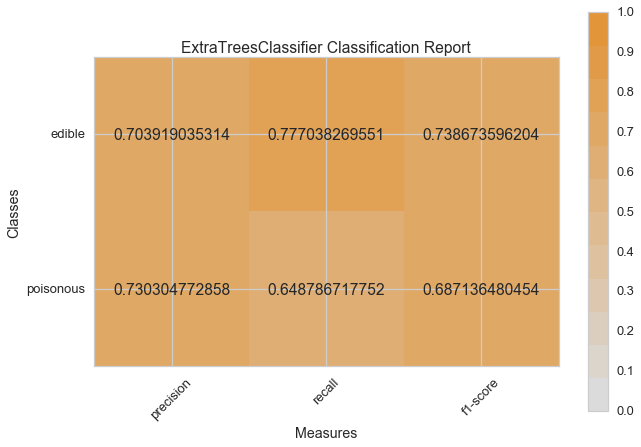
visual_model_selection(X, y, RandomForestClassifier())
Reflection¶
- Which model seems best now? Why?
- Which is most likely to save your life?
- How is the visual model evaluation experience different from numeric model evaluation?