Getting Started¶
undaqTools uses the HDF5 (Hierarchical Data Format) data model to store the data contained within NAD’s DAQ files.
undaq.py¶
The undaq.py script allows you to parallel batch processing DAQ files to HDF5. Once converted Daq instances can be loaded from HDF5 in a fraction of the time. Upon installation undaq.py should create a link in the Scripts directory of your Python installation. If your Scripts directory is in your path you should be able to call it from the command line
Note
With Windows Vista/7/8 holding down shift while right-clicking in an explorer window will add a “Open Command Window Here” option to the context menu.
Ussage is pretty straight forward. You give it a wild card argument and it searches for daq files that meet that wildcard. For example you wish to convert all daq files in the current directory:
$ undaq.py *
If each participant has there own folder from parent folder you could specify:
$ undaq.py */*
By default undaq will only run with 1 cpu. If you wish to convert daqs in parallel use the -n or –numcpu flag:
$ undaq.py */* -n 6
This would tell undaq to convert all the daq files one level down using 6 cores.
Warning
Keep in mind you might run out of memory before you run out of CPU cores. Even the fastest computers don’t work very well when they run out of memory!
By default undaq.py will only convert daq files that haven’t already been converted to hdf5. The idea being that you will probably be incrementally looking at the data as you accumulate participants. If you need to rebuild all of the hdf5 files you can use the -r or –rebuild flag.:
$ undaq.py */* -n 6 -r
if you want to output mat files for ndaqTools you can use -o mat or –output mat:
$ undaq.py */* -n 6 -o mat
By default undaq will process all the dynamic objects in the drive. If you wish to supress this use the -p or -process_dynobjs. The presence of the flag turns the processing off.
lastly there is a flag print debugging information -d or –debug:
$ undaq.py */* -n 6 -d
There are multiple level of error checking so the batch processing should never completely fail (knock-on-wood, don’t forget the caveat about memory ussage), but sometimes warnings might get thrown if the daq is missing frames or ends abruptly. When these things occur undaq does its best to fix what went wrong fix the Daq files.
Example Output:
C:\LocalData\Left Lane>undaq.py */* -n 6 -r -d
Glob Summary
--------------------------------------------------------------------
hdf5
daq size (KB) exists
--------------------------------------------------------------------
Part01\Left_01_20130424102744.daq 1,535,587 True
Part02\Left_02_20130425084730.daq 1,543,370 True
Part03\Left_03_20130425102301.daq 1,518,779 True
Part04\Left_04_20130425142804.daq 1,387,550 True
Part05\Left_05_20130425161122.daq 1,609,689 True
Part06\Left_06_20130426111502.daq 1,364,879 True
Part07\Left_07_20130426143846.daq 1,509,513 True
Part08\Left_08_20130426164114.daq 4,565 True
Part08\Left_08_20130426164301.daq 1,426,507 True
Part09\Left09_20130423155149.daq 1,339,437 True
Part10\Left10_20130423155149.daq 1,339,437 True
Part111\Left_11_20130430081052.daq 1,463,012 True
Part12\Left_12_20130429163745.daq 1,431,765 True
Part13\Left_13_20130429182923.daq 1,507,542 True
Part14\Left_14_20130430102504.daq 1,502,793 True
Part15\Left_15_20130430171947.daq 1,669,443 True
Part16\Left_16_20130501103917.daq 1,341,214 True
Part170\Left_17_20130501163745.daq 1,552,098 True
Part18\Left_18_20130502084422.daq 416,873 True
Part18\Left_18_reset_20130502090909.daq 1,128,833 True
Part19\Left_19_20130502153547.daq 1,526,510 True
Part200\Left_20_20130509094509.daq 1,572,357 True
--------------------------------------------------------------------
debug = True
rebuild = True
process_dynobjs = True
Converting daqs with 6 cpus (this may take awhile)...
Part01\Left_01_20130424102744.daq -> .hdf5 (200.7 s)
Part02\Left_02_20130425084730.daq -> .hdf5 (207.0 s)
Part03\Left_03_20130425102301.daq -> .hdf5 (193.0 s)
Part04\Left_04_20130425142804.daq -> .hdf5 (180.0 s)
Part05\Left_05_20130425161122.daq -> .hdf5 (213.1 s)
Part06\Left_06_20130426111502.daq -> .hdf5 (194.9 s)
Part07\Left_07_20130426143846.daq -> .hdf5 (194.7 s)
Part08\Left_08_20130426164114.daq -> .hdf5 (0.6 s)
Part08\Left_08_20130426164301.daq -> .hdf5 (189.3 s)
Part09\Left09_20130423155149.daq -> .hdf5 (163.3 s)
Part10\Left10_20130423155149.daq -> .hdf5 (174.0 s)
Part111\Left_11_20130430081052.daq -> .hdf5 (189.4 s)
Part12\Left_12_20130429163745.daq -> .hdf5 (193.7 s)
Part13\Left_13_20130429182923.daq -> .hdf5 (182.5 s)
Part14\Left_14_20130430102504.daq -> .hdf5 (183.9 s)
Part15\Left_15_20130430171947.daq -> .hdf5 (212.5 s)
Part16\Left_16_20130501103917.daq -> .hdf5 (173.6 s)
Part170\Left_17_20130501163745.daq -> .hdf5 (196.5 s)
Part18\Left_18_20130502084422.daq -> .hdf5 (55.2 s)
Part18\Left_18_reset_20130502090909.daq -> .hdf5 (144.5 s)
Part19\Left_19_20130502153547.daq -> .hdf5 (150.7 s)
Part200\Left_20_20130509094509.daq -> .hdf5 (152.1 s)
Debug Summary
Part01\Left_01_20130424102744.daq
Warning: Missing 3 frames. (interpolated missing frames)
Part02\Left_02_20130425084730.daq
Warning: Missing 2 frames. (interpolated missing frames)
Part06\Left_06_20130426111502.daq
Warning: Missing 1 frames. (interpolated missing frames)
Part07\Left_07_20130426143846.daq
Warning: Missing 2 frames. (interpolated missing frames)
Part08\Left_08_20130426164301.daq
Warning: Missing 1 frames. (interpolated missing frames)
Part200\Left_20_20130509094509.daq
Warning: Missing 3 frames. (interpolated missing frames)
Batch processing completed.
--------------------------------------------------------------------
Conversion Summary
--------------------------------------------------------------------
Total elapsed time: 709.3 s
Data converted: 28,995.864 MB
Data throughput: 40.9 MB/s
--------------------------------------------------------------------
C:\LocalData\Left Lane>
The Glob Summary is emitted immediately and allows users to see what files undaq has found and see whether it already has a cooresponding daq or hdf5 file converted.
After the summary undaq will tell you whether it is in debug mode, as well as whether it is in rebuild mode.
undaq.py will produce a running log alerting users as their daqs finish. The multiprocessing maintains the order from the glob summary so the output may be delayed until other files in its multiprocessing cohort complete.
To make the output more readable undaq catches and supresses the warnings until the batch processing has finished, and will only display if in debug mode.
Lastly, a conversion summary is provided. From this we can see it processed almost 30 Gb of data in 709.3 seconds at a rate of 40.9 MB/s. These particular files have the additional overhead of several hundred Ados that have to be unpacked.
Initializing and Loading Daq Instances¶
Daq objects can be initialized in two ways. The first is to read the DAQ files directly.
>>> from undaqTools import Daq
>>> daq = Daq()
>>> daq.read(daq_file)
Daq.read will also unpack and process any dynamic objects that might be present during the drive. This processing is computationally intensive can significantly increase the processing time depedending on how many dynamic objects were present. This can be suppressed with the process_dynobjs keyword argument if so desired.
>>> daq.read(daq_file, process_dynobjs=False)
Once loaded, saving the hdf5 is as simple as:
>>> daq.write_hd5(hd5_file)
Assuming you have already converted your DAQS to HDF5 using the above method or using the undaq.py script you could load the HDF5 file directly:
>>> daq.read_hd5(hd5_file)
Reading HDF5 files is about 2 magnitude orders faster than reading DAQ files directly. The suggested interaction is to use the undaq.py script to convert your DAQ files to HDF5. Once they have been converted the DAQs should be archived but should not be needed for the data analysis.
Specifying an elemlist¶
To save memory and time a list of element wildcards to load can be supplied to read_hd5:
>>> elemlist = ['VDS_Veh*', 'CFS_Accelerator_Pedal_Position']
>>> daq.read_hd5(hd5_file, elemlist=elemlist)
Every wildcard in elemlist is matched to the elements in the HDF5 file. The above example would load all the elements beginning with ‘VDS_Veh’ and ‘CFS_Accelerator_Pedal_Position’.
The wildcard matching is Unix-shell style case-normalized matching (i.e. not regex).
Note
No warnings are provided if your wildcards don’t match the elements in the HDF5.
Daq.match_keys()¶
The DAQ files provide an almost overwhelming amount of data. When you first start getting acquainted with your driving simulator data it is easy to forget what contain the the things that you are interested in. The match_keys function makes this a little easier by allowing you to find keys that match Unix style wildcard patterns. The searches are case insensitive.
>>> daq.match_keys('*veh*dist*')
[u'VDS_Veh_Dist', u'SCC_OwnVeh_PathDist', u'SCC_OwnVehToLeadObjDist']
stat() and the Daq.info <NamedTuple>¶
If you want to get metadata from a DAQ file but don’t have it converted to HDF5 you can use the undaqTools.stat() function to pull out the info metadata.
>>> from undaqTools import stat
>>> info = stat('data reduction_20130204125617.daq')
Info(run='data reduction',
runinst='20130204125617',
title='Nads MiniSim',
numentries=245,
frequency=59,
date='Mon Feb 04 12:56:17 2013\n',
magic='7f4e3d2c',
subject='part12',
filename='data reduction_20130204125617.daq')
When you load data into a Daq instance the info attribute contains this metadata. The Info dict is stored as a namedTuple to prevent or at least discourage users from altering this metadata.
Daq.etc <dict>¶
The data reductions are usually hypothesis driven. This means that we need to obtain dependent measures reflecting the conditions of independent variables. To perform the statistical analyses we need to keep track of these things as well as other metadata. Every Daq instance has an etc dictionary that can be used to store this metadata. Daq.write_hd5() will export the etc dict and Daq.read_hd5() will restore it.
>>> daq.etc['Gender'] = 'M'
>>> daq.etc['Factor1'] = [ 10, 20, 10, 20, 10, 20]
>>> daq.etc['Factor2'] = ['A','A','A','B','B','B']
The hdf5 file format is somewhat limited in the datatypes that it can store. To get data in and out of hdf5 repr is applied to the values and a modified version of ast.literal_eval is used to get the data back out. this is to avoid security vulnerabilities with untrusted hdf5 Daq representations (if eval was used). As a result only Python literal structures (strings, numbers, tuples, lists, dicts, booleans, and None) and undaqTools FrameSlice and FrameIndex objects can be stored. The code doesn’t check to see if the etc dict will export and import when the attribute changes. To get this to work you would have to interact with etc through getter and setter methods and it just doesn’t seem particularly worth the hassle.
Accessing Element Data¶
Daq objects are dictionary objects. The keys coorespond to the NADS variable names in the DAQ files. The values are Element object instances. The Element class inherents numpy ndarrays and they are always 2 dimensional.
>>> daq['VDS_Veh_Speed'].shape
(1L, 10658L)
Because Element is a numpy.ndarray subclass they behave, for the most part, just like the plain old numpy arrays that you are (hopefully) use to.
>>> np.mean(daq['VDS_Veh_Speed'])
76.4363
They also keep track of the frames that their data represent. The frames are always a 1 dimensional and are aligned with the second axis of the Element’s data.
>>> veh_spd = daq['VDS_Veh_Speed']
>>> type(veh_spd.frames)
<type 'numpy.ndarray'>
>>> veh_spd.shape
(10658L,)
Dynamic objects also contain attribute data as Elements and may only be present during a subset of the drive. Because the dynamic object data and the CSSDC measures are unaligned with the Elements it is not always possible or convenient to simply use indexes to slice Elements. We need to slice based on frames. This is possible with fslice()
>>> daq['VDS_Veh_Speed'][0, fslice(4000, 4010)]
Element(data = [ 42.17745972 42.3068924 42.4354744 42.56311417 42.68973923
42.81529999 42.93975449 43.06305313 43.18511963 43.3058815 ],
frames = [4000 4001 4002 4003 4004 4005 4006 4007 4008 4009],
name = 'VDS_Veh_Speed',
numvalues = 1,
rate = 1,
varrateflag = False,
nptype = float32)
As the reader can see from the string representation other metadata from the header block of the DAQ file gets attached to the Element.
CSSDC Elements¶
Many of the available measures are Change State Signal Detection (CSSDC) measures. they contains categorical data that only updates when a change in state is detected.
>>> daq['TPR_Tire_Surf_Type']
Element(data = [[11 1 1 11 11 11 1 1 11 11 3 3 3 3 3 3 11 11 1 1 11 11 1 1]
[11 1 1 11 11 11 1 1 11 11 11 11 3 3 11 11 11 11 1 1 11 11 1 1]
[11 11 1 1 1 11 11 1 1 11 11 3 3 3 3 3 3 11 11 1 1 11 11 1]
[11 11 1 1 11 11 11 1 1 11 11 11 11 3 3 11 11 11 11 1 1 11 11 1]
[ 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0]
[ 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0]
[ 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0]
[ 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0]
[ 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0]
[ 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0]],
frames = [ 2716 5519 5523 5841 5844 5845 7970 7973 8279 8284 8785 8791
8818 8824 9127 9132 9166 9171 10270 10274 10597 10600 12655 12659],
name = 'TPR_Tire_Surf_Type',
numvalues = 10,
rate = -1 (CSSDC),
varrateflag = False,
nptype = int16)
The above example contains data pertaining to surface type for the 4 tires and has 6 unfilled rows for additional tires.
All elements with a rate != 1 (as defined in the DAQ file) are considered CSSDC. We can check this with isCSSDC()
>>> daq['TPR_Tire_Surf_Type'].isCSSDC()
True
>>> daq['VDS_Veh_Speed'].isCSSDC()
False
Use findex() to get the state at a given frame (even if the frame is not defined)
>>> # frame 5800 is not explictly defined
>>> daq['TPR_Tire_Surf_Type'][:4, findex(5800)]
array( [[ 1],
[ 1],
[ 1],
[ 1]], dtype=np.int16)
If you ask for a frame before the first defined frame you will get nan. If you ask for a frame after the last defined frame you will get the last frame.
method it is easy to test whether an Element contains CSSDC data. The value at any frame between the first and last frame defined for a CSSDC Element can be obtained through slicing. This treats the data as categorical and always returns the last defined state.
Timeseries Plots¶
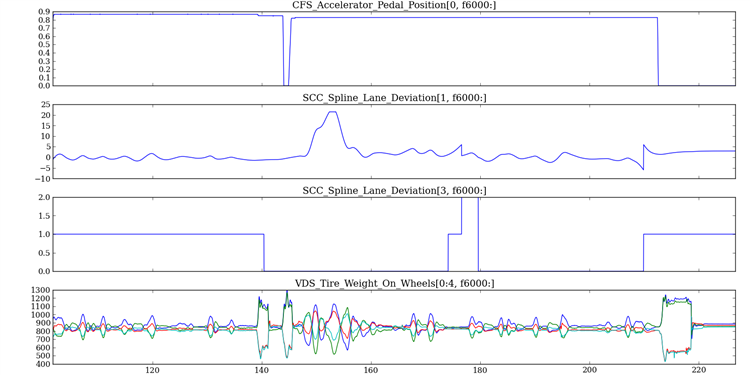
The Daq Class has some built-in visualization routines. Multipanel timeseries plots can be constructed with the plot_ts() method. The method takes a list of tuples containing the element names and row indices to plot. Each list argument becomes a subplot. The xindx keyword allows one to control the range of the x-axis across all of the subplots.
The code is smart enough to dynamically adjust its height as additional subplots are specified. It also knows to represent CSSDC measures as step functions. The method returns a matplotlib.figure.Figure instance.
Building a timeseries plot:
import matplotlib.pyplot as plt
from undaqTools import Daq
elems_indxs = [('CFS_Accelerator_Pedal_Position', 0),
('SCC_Spline_Lane_Deviation', 1),
('SCC_Spline_Lane_Deviation_Fixed', 0),
('SCC_Spline_Lane_Deviation', 3),
('VDS_Tire_Weight_On_Wheels', slice(0,4))]
daq = Daq()
daq.read_hd5(hdf5file)
fig = daq.plot_ts(elems_indxs, xindx=fslice(6000, None))
fig.savefig('ts_plot.png')
plt.close('all')
Download [hi-res]
DynObjs Plots¶
The Daq Class also has a routine to visualize the pathes taken by the OwnVehicle and dynamic objects.
dynobjs plot:
import matplotlib.pyplot as plt
from undaqTools import Daq
daq = Daq()
daq.read_hd5(hdf5file)
fig = daq.plot_dynobjs('Ado*')
fig.savefig('dynobjs_plot.png')
plt.close('all')
Download [hi-res]

Quirks¶
Absence of Time
Time is almost completly redundant with the frames data for most things. Just start thinking in frames. It will soon become second nature.
If you do need millisecond precision timing there is the ‘SCC_Graphics_Wall_Clock_Time’ element. It an int32 Element with millisecond units. It doesn’t start at 0.