biobase: base structures and functions for Bioconductor¶
The module reflects the content of the R/Bioconductor package Biobase. It defines Python-level classes for the R/S4 classes, and gives otherwise access to R-level commands the usual rpy2:robjects way.
The variable biobase_env in the module is an rpy2.robjects.REnvironment for the modules namespace. Accessing explicitly a module’s object is then straightforward.
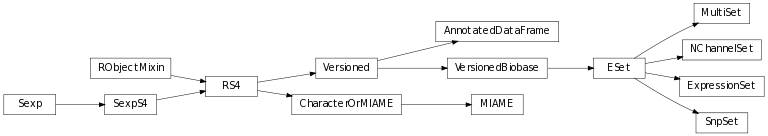
The class inheritance diagram presents the parent-child relationships between the elements.
A module to model the Biobase library in Bioconductor
Copyright 2009-2010 - Laurent Gautier
- class bioc.biobase.AnnotatedDataFrame¶
An annotated data.frame as defined in the R package Biobase
- combine(annotated_dataframe)¶
- Combine the instance with an other data.frame
- featurenames¶
- Property for both R’s ‘featureNames’ and ‘featureNames<-‘
- get_featurenames()¶
- get_pdata()¶
- classmethod new(data=<DataFrame - Python:0x39355a8 / R:0x2c39000>, varmetadata=<DataFrame - Python:0x3935580 / R:0x2c35774>, dimlabels=<StrVector - Python:0x3935558 / R:0x2e55e28>)¶
- pdata¶
- Property for both R’s ‘pData’ and ‘pData<-‘
- set_featurenames(value)¶
- set_pdata(value)¶
- class bioc.biobase.CharacterOrMIAME¶
- Abstract class
- class bioc.biobase.ESet¶
An eSet as defined in the R package Biobase. This class in defined as “virtual” in the R/S4 scheme, which can translates as “abstract” in more common OO terminologies.
- annotation()¶
- assaydata()¶
- experimentdata()¶
- get_featuredata()¶
- get_phenodata()¶
- set_featuredata(value)¶
- set_phenodata(value)¶
- class bioc.biobase.ExpressionSet(*args, **kwargs)¶
- esapply(margin, fun, *args)¶
- “apply” a function “fun” along the “margin”.
- exprs¶
- Property for both R’s ‘exprs’ and ‘exprs<-‘
- get_exprs()¶
- classmethod new(phenodata=<RS4 - Python:0x390edf0 / R:0x30754f4>, featuredata=<RS4 - Python:0x3935da0 / R:0x2df612c>, experimentdata=<RS4 - Python:0x39354b8 / R:0x30d9dd4>, annotation=<StrVector - Python:0x3935170 / R:0x30db8cc>, exprs=<Matrix - Python:0x39357b0 / R:0x2f7b4a8>)¶
- set_exprs(value)¶
- class bioc.biobase.MIAME¶
- abstract¶
- maps Biobase::abstract
- expinfo¶
- maps Biobase::expinfo
- hybridizations¶
- maps Biobase::hybridizations
- normcontrols¶
- maps Biobase::normControls
- notes¶
- maps Biobase::notes
- otherinfo¶
- maps Biobase::otherInfo
- preproc¶
- maps Biobase::preproc
- samples¶
- maps Biobase::samples
- class bioc.biobase.MultiSet¶
- class bioc.biobase.NChannelSet¶
- channel(name, **kwargs)¶
- channelnames¶
- maps Biobase::channelNames
- selectchannels(names, **kwargs)¶
- class bioc.biobase.SnpSet¶
- class bioc.biobase.Versioned¶
- class bioc.biobase.VersionedBiobase¶
- bioc.biobase.biobase_conversion(robj)¶