Fitting of Experimental Data¶
Introduction¶
These routines / classes provide a method for fitting of data using mostly least squares methods. There are two main methods here. The pyspec.fit.fit class provides a class for fitting of data. The pyspec.fit.fitdata subroutine serves as a wrapper around the pyspec.fit.fit class. Here the data is taken from the current selected figure. Basically this means that you can fit any set of data which is on the current figure.
The fit Class¶
Fitting user supplied data¶
- class pyspec.fit.fit(x=None, y=None, e=None, funcs=None, guess=None, quiet=False, optimizer='mpfit', ifix=None, ilimited=None, ilimits=None, xlimits=None, xlimitstype='world', interactive=False, r2min=0.0, debug=0)¶
General class to perform non-linear least squares fitting of data.
This class serves as a wrapper around various fit methods, requiring a standard function to allow for easy fitting of data.
- x : ndarray
- x data
- y : ndarray
- y data
- e : ndarray
- y error
- funcs : list
- A list of functions to be fitted. These functions should be formed like the standard fitting functions in the pyspec.fitfuncs.
- guess : array or string
- Array of the initial guess, or string for guess type:
- ‘auto’ : Auto guess of parameters
- quiet : bool
- If True then don’t print to the console any results.
- ifix : ndarray
- An array containing a ‘1’ for fixed parameters
- xlimits : ndarray
- An (n x 2) array of the limits in x to fit.
- xlimitstype : string
- Either ‘world’ or ‘index’
- optimiser : string :
- Optimizer to use for fit: ‘mpfit’, ‘leastsq’, ‘ODR’
- interactive : bool
- True/False launch interactive fitting mode
- r2min : float
- If r^2 is less than this value, drop into interactive mode
- After a sucsesful fit, the class will contain members for the following.
- {fit}.result
- result of fit
- {fit}.stdev
- errors on results (sigma)
- {fit}.fit_result
- an array of both results and stdev for convinience
- {fit}.covar
- covarience matrix of result
- {fit}.corr
- correlation matrix of result
- {fit}.r2
- R^2 value for fit.
- chiSquared(norm=True, dist='poisson')¶
Return the chi-squared value for the fit
- Calculate the chi-squared value for the fit. This is defined as
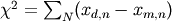
- Where d is the data and m is the model. The normalized chi-squared is given by
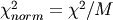
where
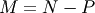 , where P is the number of parameters.
, where P is the number of parameters.If dist is ‘poisson’ then the data is divided by the model answer. i.e.
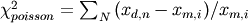
- norm : bool
- If true calculate the normalized chi-squared.
- dist : string
- The distribution to use, currently only ‘poisson’
chi-squared value
- evalfitfunc(nxpts=None, p=None, x=None)¶
Evaluate the fit functions with the fesult of a fit
- nxpts : int
- Number of x data points if using the range of the input data. If none then the x points of the dataset are used.
- p : ndarray
- Parameters of function. If None, use current fit result.
- x : ndarray
- Evaluate fit function at each point defined by the ndarray.
returns f(x) : ndarray
- go(interactive=False)¶
Start the fit
- interactive : bool
- If True, start the fit in interactive mode.
- residualSDev()¶
Calculate the sandard deviation of the residuals
- run(interactive=False)¶
Start the fit
- interactive : bool
- If True, start the fit in interactive mode.
- textResult()¶
Return a string containing the text results of the fit
Fitting data plotted using matplotlib¶
- pyspec.fit.fitdata(funcs, ax=None, showguess=False, *args, **kwargs)¶
Force fitting of a function to graphed data
- funcs : list
- list of the fit functions
- ax : matplotlib axis instance
- axis to fit (if none the current axis is used)
All the additional *args and **kwargs are passed onto the fit class (see class pyspec.fit.fit for details).
pyspec.fit.fit instance with result.
Standard Fit Functions¶
- pyspec.fitfuncs.constant(x, p, mode='eval')¶
Single constant value
- Function:
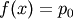
- pyspec.fitfuncs.gauss(x, p, mode='eval')¶
Gaussian defined by amplitide
- Function:
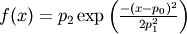
- pyspec.fitfuncs.linear(x, p, mode='eval')¶
Linear (strait line)
- Function:
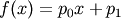
- pyspec.fitfuncs.lor(x, p, mode='eval')¶
Lorentzian defined by amplitude
- Function:
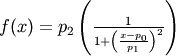
- pyspec.fitfuncs.lor2(x, p, mode='eval')¶
Lorentzian squared defined by amplitude
- Function:
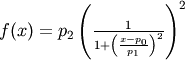
- The HWHM is related to the parameter
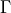 by the relation:
by the relation: 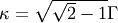
- pyspec.fitfuncs.lor2a(x, p, mode='eval')¶
Lorentzian squared function defined by area
- pyspec.fitfuncs.lorr(x, p, mode='eval')¶
Lorentzian defined by area
- Function:
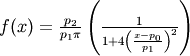
- pyspec.fitfuncs.peakguess(x, y)¶
Guess the “vital statistics” of a peak.
This function guesses the peak’s center width, integral, height and linear background (m, c) from the x and y data passed to it.
- pyspec.fitfuncs.power(x, p, mode='eval')¶
Power function
- Function:
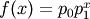
- pyspec.fitfuncs.pvoight(x, p, mode='eval')¶
Pseudo Voight function
- Function:
- :math:’f(x) = ‘
User Defined Functions¶
User defined functions can easily be written. An example of a fit function to provide a linear (straight line) is shown below:
def linear(x, p, mode='eval'):
if mode == 'eval':
out = (p[0] * x) + p[1]
elif mode == 'params':
out = ['grad','offset']
elif mode == 'name':
out = "Linear"
elif mode == 'guess':
g = peakguess(x, p)
out = g[4:6]
else:
out = []
return out
This function is called by the fitting routine each time the function needs to be evaluated. It is also called other times, to gain information about the function. The mode parameter is used to let the function know what is required. The default value of mode should be set to 'eval'. The possible values of mode are discussed below
- eval
- Evaluate function. Here the function should return
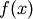 from x and p, where p is the parameters
from x and p, where p is the parameters - params
- List of parameter names. Here the function should return a list of strings describing the fit parameters. This list should have the same length as the number of fit parameters.
- name
- Name of the function. Here the function should return a string with a text description of the function.
- guess
- Parameter guess. Here the function should return a guess of
the fit parameters. The function is passed
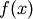 through
the second parameter p. The function should return it’s best
guess based on the experimental data, and return those
parameters. The peakguess function is provided for convenience
which can guess the vital statistics of a peak from the
data. This option is not essential but if it is not implemented
then the user must provide a numerical guess to the fit class.
through
the second parameter p. The function should return it’s best
guess based on the experimental data, and return those
parameters. The peakguess function is provided for convenience
which can guess the vital statistics of a peak from the
data. This option is not essential but if it is not implemented
then the user must provide a numerical guess to the fit class.
Finally, the default response should be to return a null list [].