glimpse.glab.api¶
- DEFAULT_LAYER¶
Default model layer to use for evaluation.
- SetParamsWithGui(params=None, **kw)[source]¶
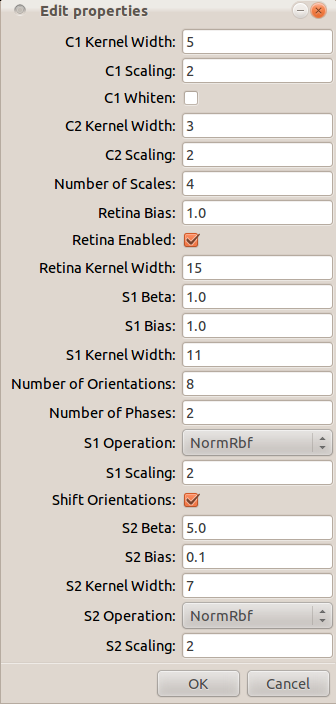
Choose model parameters using a graphical interface.
This presents a display similar to that shown in Figure 1.
- GetExperiment()[source]¶
Get the current experiment object.
This is an advanced function. In general, the user should not modify the experiment object directly.
- Verbose(flag=True)[source]¶
Set the verbosity of log output.
Parameters: flag (bool) – Whether to enable verbose logging.
- GetModel()[source]¶
Get the Glimpse model used for this experiment.
This is an advanced function. In general, the user should not need to interact with the model directly.
- SetCorpus(corpus_dir, balance=False)[source]¶
Read images from the corpus directory.
This function assumes that each sub-directory contains images for exactly one object class, with a different object class for each sub-directory. Training and testing subsets are chosen automatically.
Parameters: - root_dir (str) – Path to corpus directory.
- balance (bool) – Ensure an equal number of images from each class (by random selection).
- reader – Filesystem reader.
See also
- SetCorpusSubdirs(corpus_subdirs, balance=False)[source]¶
Read images from per-class corpus sub-directories.
This function assumes that each sub-directory contains images for exactly one object class, with a different object class for each sub-directory. Training and testing subsets are chosen automatically.
Parameters: - subdirs (iterable of str) – Path of each corpus sub-directory.
- balance (bool) – Ensure an equal number of images from each class (by random selection).
- reader – Filesystem reader.
See also
- SetCorpusSplit(train_dir, test_dir)[source]¶
Read images and training information from the corpus directory.
This function assumes that the train_dir and test_dir have the same set of sub-directories. Each sub-directory shoudl contain images for exactly one object class, with a different object class for each sub-directory.
Parameters: - train_dir (str) – Path to corpus of training images.
- test_dir (str) – Path to corpus of test images.
- reader – Filesystem reader.
See also
- SetCorpusByName(name)[source]¶
Use a sample image corpus for this experiment.
Parameters: name (str) – Corpus name. One of ‘easy’, ‘moderate’, or ‘hard’. This provides access to a small set of images for demonstration purposes, which are composed of simple shapes on various background patterns.
- SetS2Prototypes(prototypes)[source]¶
Manually specify the set of S2 prototypes.
Parameters: prototypes (str or list of array of float) – Path to prototypes on disk, or prototypes array to set. Return type: list of array of float Returns: Set of model prototypes.
- ImprintS2Prototypes(num_prototypes)[source]¶
Create a set of S2 prototypes by “imprinting” from training images.
Patches are drawn from all classes of the training data.
Parameters: num_prototypes (int) – Number of prototypes to create.
- MakeUniformRandomS2Prototypes(num_prototypes, low=None, high=None)[source]¶
Create a set of random S2 prototypes drawn from the uniform distribution.
Each element of every prototype is drawn independently from an uniform distribution with the same parameters.
Parameters: - num_prototypes (int) – Number of prototypes to create.
- low (float) – Minimum value in uniform range.
- high (float) – Maximum value in uniform range.
- MakeShuffledRandomS2Prototypes(num_prototypes)[source]¶
Create a set of “imprinted” S2 prototypes that have been shuffled.
Each prototype has its contents randomly permuted across location and orientation band.
Parameters: num_prototypes (int) – Number of prototypes to create.
- MakeHistogramRandomS2Prototypes(num_prototypes)[source]¶
Create a set of S2 prototypes drawn from a 1D histogram of C1 activity.
The set is created by drawing elements from a distribution that is estimated from a set of imprinted prototypes. Each entry is drawn independently of the others.
Parameters: num_prototypes (int) – Number of prototypes to create.
- MakeNormalRandomS2Prototypes(num_prototypes)[source]¶
Create a set of random S2 prototypes drawn from the normal distribution.
Each element of every prototype is drawn independently from a normal distribution with the same parameters.
Parameters: num_prototypes (int) – Number of prototypes to create.
- MakeKmeansS2Prototypes(num_prototypes, num_patches=None)[source]¶
Create a set of S2 prototypes by clustering C1 samples with k-Means.
Parameters: - num_prototypes (int) – Number of prototypes to create.
- num_patches (int) – Number of sample patches passed to k-Means.
- ComputeActivation(save_all=False)[source]¶
Compute the model activity for all images in the experiment.
Parameters: - layers (str or list of str) – One or more model layers to compute.
- pool – Worker pool to use for parallel computation.
- save_all (bool) – Whether to save activation for all model layers, rather than just those in layers.
- progress – Handler for incremental progress updates.
- EvaluateClassifier(cross_validate=False, algorithm=None, train_size=None, num_folds=None, score_func=None)[source]¶
Apply a classifier to the image features in the experiment.
Parameters: - cross_validate (bool) – Whether to use cross-validation. The default will use a fixed training and testing split.
- algorithm – Learning algorithm, which is fit to features. This should be a scikit-learn classifier object. If not set, a LinearSVC object is used.
- train_size (float or int) – Size of training split, specified as a fraction (between 0 and 1) of total instances or as a number of instances (1 to N, where N is the number of available instances).
- num_folds (int) – Number of folds to use for cross-validation. Default is 10.
- score_func (str) – Name of the scoring function to use, as specified by ResolveScoreFunction().
Return type: ExperimentData
Returns: Results of evaluation.
- GetNumPrototypes(kwidth=0)[source]¶
Return the number of S2 prototypes in the model.
Parameters: kwidth (int) – Index of kernel shape.
- GetPrototype(prototype=0, kwidth=0)[source]¶
Return an S2 prototype from the experiment.
Parameters: - prototype (int) – Index of S2 prototype.
- kwidth (int) – Index of kernel shape.
- GetImprintLocation(prototype=0, kwidth=0)[source]¶
Return the image location from which a prototype was imprinted.
This requires that the prototypes were learned by imprinting.
Parameters: - prototype (int) – Index of S2 prototype.
- kwidth (int) – Index of kernel shape.
Returns: Location information in the format (image index, scale, y-offset, x-offset), where scale and y- and x-offsets identify the S2 unit from which the prototype was “imprinted”.
Return type: 4 element array of int
- GetEvaluationLayers(evaluation=0)[source]¶
Returns the model layers from which features were extracted.
Parameters: evaluation (int) – Index of the evaluation record to use. Return type: list of str Returns: Names of layers used for evaluation.
- GetEvaluationResults(evaluation=0)[source]¶
Returns the results of a model evaluation.
Parameters: evaluation (int) – Index of the evaluation record to use. Return type: glimpse.util.data.Data Returns: Result data, with attributes that depend on the method of evaluation. In general, the feature_builder, score, score_func attributes will be available.
- GetPredictions(training=False, evaluation=0)[source]¶
Get information about classifier predictions.
Parameters: - exp – Experiment data.
- training (bool) – Return information about training images. Otherwise, information about the test set is returned.
- evaluation (int) – Index of the evaluation record to use.
Return type: list of 3-tuple of str
Returns: filename, true label, and predicted label for each image in the set
- ShowS2Activity(image=0, scale=0, prototype=0, kwidth=0)[source]¶
Plot the S2 activity for a given image.
Parameters: - image – Path to image on disk, or index of image in experiment.
- scale (int) – Index of scale band to use.
- prototype (int) – Index of S2 prototype to use.
- kwidth (int) – Index of kernel shape.
- ShowPrototype(prototype=0, kwidth=0)[source]¶
Plot the prototype activation.
There is one plot for each orientation band.
Parameters: - prototype (int) – Index of S2 prototype to use.
- kwidth (int) – Index of kernel shape.
- colorbar (bool) – Add a colorbar to the plot.
- AnnotateImprintedPrototype(prototype=0, kwidth=0)[source]¶
Plot the image region used to construct a given imprinted prototype.
This shows the image in the background, with a red box over the imprinted region.
Parameters: - prototype (int) – Index of S2 prototype to use.
- kwidth (int) – Index of kernel shape.
- AnnotateS2Activity(image=0, scale=0, prototype=0, kwidth=0)[source]¶
Plot the S2 activity and image data for a given image.
This shows the image in the background, with the S2 activity on top.
Parameters: - image – Path to image on disk, or index of image in experiment.
- scale (int) – Index of scale band to use.
- prototype (int) – Index of S2 prototype to use.
- kwidth (int) – Index of kernel shape.
- AnnotateC1Activity(image=0, scale=0)[source]¶
Plot the C1 activation for a given image.
This shows the image in the background, with the activation plotted on top. There is one plot for each orientation band.
Parameters: - image – Path to image on disk, or index of image in experiment.
- scale (int) – Index of scale band to use.
- AnnotateS1Activity(image=0, scale=0)[source]¶
Plot the S1 activation for a given image.
This shows the image in the background, with the activation plotted on top. There is one plot for each orientation band.
Parameters: - image – Path to image on disk, or index of image in experiment.
- scale (int) – Index of scale band to use.
- colorbar (bool) – Whether to show colorbar with plot.