Valve manual¶
Valve application is a driver that uses aquaduct module to perform analysis of trajectories of selected residues in Molecular Dynamics simulation.
Valve invocation¶
Once aquaduct module is installed (see Aqua-Duct installation guide) properly on the machine Valve is available as valve.py command line tool.
Usage¶
Basic help of Valve usage can be displayed by following command:
valve.py --help
It should display following information:
usage: valve.py [-h] [--debug] [--debug-file DEBUG_FILE]
[--dump-template-config] [-t THREADS] [-c CONFIG_FILE] [--sps]
[--max-frame MAX_FRAME] [--min-frame MIN_FRAME]
[--step-frame STEP_FRAME] [--version] [--license]
Valve, Aquaduct driver
optional arguments:
-h, --help show this help message and exit
--debug Prints debug info. (default: False)
--debug-file DEBUG_FILE
Debug log file. (default: None)
--dump-template-config
Dumps template config file. Suppress all other output
or actions. (default: False)
-t THREADS Limit Aqua-Duct calculations to given number of
threads. (default: None)
-c CONFIG_FILE Config file filename. (default: None)
--sps Use single precision to store data. (default: False)
--max-frame MAX_FRAME
Maximal number of frame. (default: None)
--min-frame MIN_FRAME
Minimal number of frame. (default: None)
--step-frame STEP_FRAME
Frames step. (default: None)
--version Prints versions and exits. (default: False)
--license Prints short license info and exits. (default: False)
Configuration file template¶
Configuration file used by Valve is of moderate length and complexity. It can be easily prepared with a template file that can be printed by Valve. Use following command to print configuration file template on the screen:
valve.py --dump-template-config
Configuration file template can also be easily saved in to a file with:
valve.py --dump-template-config > config.txt
Where config.txt is a configuration file template.
For detailed description of configuration file and available options see Configuration file options.
Valve calculation run¶
Once configuration file is ready Valve calculations can be run with a following simple command:
valve.py -c config.txt
Some of Valve calculations can be run in parallel. By default all available CPU cores are used. This is not always desired - limitation of used CPU cores can be done with -t option which limits number of concurrent threads used by Valve. If it equals 1 no parallelism is used.
Note
Specifying number of threads greater then available CPU cores is generally not optimal.
However, in order to maximize usage of available CPU power it is recommended to set it as number of cores + 1. The reason is that Valve uses one thread for the main process and the excess over one for processes for parallel calculations. When parallel calculations are executed the main thread waits for results.
Note
Options --min-frame, --max-frame, and --step-frame can be used to limit calculations to specific part of trajectory. For example, to run calculations for 1000 frames starting from frame 5000 use following options: --min-frame 4999 --max-frame 5999; to run calculations for every 5th frame use: --step-frame 5.
Single precision storage¶
Most of the calculation is Valve is performed by NumPy. By default, NumPy uses double precision floats.
Valve does not change this behavior but has special option --sps which forces to store all data (both internal data stored in RAM and on the disk) in single precision. This spare a lot of RAM and is recommended what you perform calculation for long trajectories and you have limited amount of RAM.
Debuging¶
Valve can output some debug information. Use --debug to see all debug information on the screen or use --debug-file with some file name to dump all debug messages to the given file. Beside debug messages standard messages will be saved in the file as well.
How does Valve work¶
Application starts with parsing input options. If --help or --dump-template-config options are provided appropriate messages are printed on the screen and Valve quits with signal 0.
Note
In current version Valve does not check the validity of the config file.
If config file is provided Valve parse it quickly and regular calculations starts according to its content. Calculations performed by Valve are done in several stages described in the next sections.
Traceable residues¶
In the first stage of calculation Valve finds all residues that should be traced and appends them to the list of traceable residues. It is done in a loop over all frames. In each frame residues of interest are searched and appended to the list but only if they are not already present on the list.
The search of the residues is done according to user provided definitions. Two requirements have to be met to append residue to the list:
- The residue has to be found according to the object definition.
- The residue has to be within the scope of interest.
The object definition encompasses usually the active site of the protein. The scope of interest defines, on the other hand, the boundaries in which residues are traced and is usually defined as protein.
Since aquaduct in its current version uses MDAnalysis Python module for reading, parsing and searching of MD trajectory data, definitions of object and scope have to be given as its Selection Commands.
Object definition¶
Object definition has to comprise of two elements:
- It has to define residues to trace.
- It has to define spatial boundaries of the object site.
For example, proper object definition could be following:
(resname WAT) and (sphzone 6.0 (resnum 99 or resnum 147))
It defines WAT as residues that should be traced and defines spatial constrains of the object site as spherical zone within 6 Angstroms of the center of masses of residues with number 99 and 147.
Scope definition¶
Scope can be defined in two ways: as object but with broader boundaries or as the convex hull of selected molecular object.
In the first case definition is very similar to object and it has to follow the same limitations. For example, proper scope definition could be following:
resname WAT around 2.0 protein
It consequently has to define WAT as residues of interest and defines spatial constrains: all WAT residues that are within 2 Angstroms of the protein.
If the scope is defined as the convex hull of selected molecular object (which is recommended), the definition itself have to comprise of this molecular object only, for example protein. In that case the scope is interpreted as the interior of the convex hull of atoms from the definition. Therefore, traceable residues would be in the scope only if they are within the convex hull of atoms of protein.
Convex hulls of macromolecule atoms¶
AQ uses quickhull algorithm for convex hulls calculations (via SciPy class scipy.spatial.ConvexHull, see also http://www.qhull.org/ and original publication The quickhull algorithm for convex hulls).
Convex hull concept is used to check if traced molecules are inside of the macromolecule. Convex hull can be considered as rough approximation of molecular surface. Following picture shows schematic comparison of convex hull and solvent excluded surface:
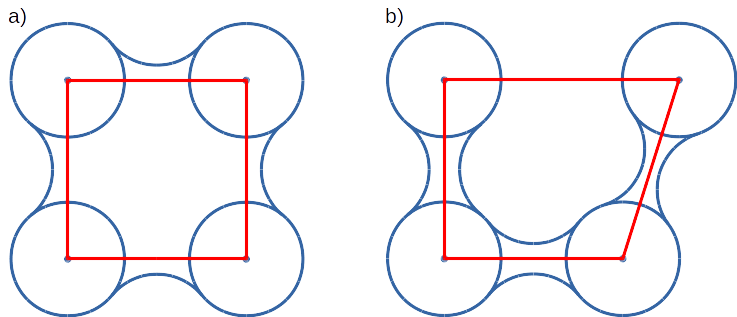
Convex hull (red shape) of atoms (blue dots with VdW spheres) and SES (blue line): a) Convex hull and SES cover roughly the same area, Convex hull approximates SES; b) movement of one atom dramatically changes SES, however, interior of the molecule as approximated by Convex hull remains stable.
No doubts, Convex hull is a very rough approximation of SES. It has, however, one very important property when it is used to approximate interior of molecules: its interior does not considerably depend on the molecular conformation of a molecule (or molecular entity) in question.
Raw paths¶
The second stage of calculations uses the list of all traceable residues from the first stage and finds coordinates of center of masses for each residue in each frame. As in the first stage, it is done in a loop over all frames. For each residue in each frame Valve calculates or checks two things:
- Is the residue in the scope (this is always calculated according to the scope definition).
- Is the residue in the object. This information is partially calculated in the first stage and can be reused in the second. However, it is also possible to recalculate this data according to the new object definition.
For each of the traceable residues a special Path object is created. If the residue is in the scope its center of mass is added to the appropriate Path object together with the information if it is in the object or not.
Separate paths¶
The third stage uses collection of Path objects to create Separate Path objects. Each Path comprise data for one residue. It may happen that the residue enters and leaves the scope and the object many times over the entire MD. Each such event is considered by Valve as a separate path.
There are two types of Separate Paths:
- Object Paths
- Passing Paths
Object Paths are traces of molecules that visited Object area. Passing Paths are traces of molecules that entered Scope but did not entered Object area.
Passing paths comprises of one part only. Each object path comprises of three parts:
- Incoming - Defined as a path that leads from the point in which residue enters the scope and enters the object for the first time.
- Object - Defined as a path that leads from the point in which residue enters the object for the first time and leaves it for the last time.
- Outgoing - Defined as a path that leads from the point in which residue leaves the object for the last time and leaves the scope.
It is also possible that incoming and/or outgoing part of the separate path is empty.
Note
Generation of Passing paths is optional and can be switched off.
Warning
Generation of Passing paths without redefinition of Object area in stage I and II may lead to false results.
Auto Barber¶
After the initial search of Separate Path objects it is possible to run procedure, Auto Barber, which trims paths down to the approximated surface of the macromolecule or other molecular entity defined by the user. This trimming is done by creating collection of spheres that have centers at the ends of paths and radii equal to the distance for the center to the nearest atom of user defined molecular entity. Next, parts of raw paths that are inside these spheres are removed and separate paths are recreated.
Auto Barber procedure has several options, for example:
- auto_barber allows to define molecular entity which is used to calculate radii of spheres used for trimming raw paths.
- auto_barber_mincut allows to define minimal radius of spheres. Spheres of radius smaller then this value are not used in trimming.
- auto_barber_maxcut allows to define maximal radius of spheres. Spheres of radius greater then this value are not used in trimming.
- auto_barber_tovdw if set to True radii of spheres are corrected (decreased) by Van der Waals radius of the closest atom.
See also options of separate_paths stage.
Smoothing¶
Separate paths can be optionally smoothed. This can be done in two modes: soft and hard. In the former mode smoothed paths are used only for visualization purposes. In the latter, raw paths are replaced by smoothed.
Note
If hard mode is used all further calculations are performed for smoothed paths.
Available methods¶
Aqua-Duct implements several smoothing methods:
- Savitzky-Golay filter -
SavgolSmooth- see also original publication Smoothing and Differentiation of Data by Simplified Least Squares Procedures (doi:10.1021/ac60214a047). - Window smoothing -
WindowSmooth - Distance Window smoothing -
DistanceWindowSmooth - Active Window smoothing -
ActiveWindowSmooth - Max Step smoothing -
MaxStepSmooth - Window over Max Step smoothing -
WindowOverMaxStepSmooth - Distance Window over Max Step smoothing -
DistanceWindowOverMaxStepSmooth - Active Window over Max Step smoothing -
ActiveWindowOverMaxStepSmooth
For detailed information on available configuration options see configuration file smooth section description.
Clusterization of inlets¶
Each of the separate paths has beginning and end. If they are at the boundaries of the scope they are considered as Inlets, i.e. points that mark where the traceable residues enters or leaves the scope. Clusters of inlets, on the other hand, mark endings of tunnels or ways in the system which was simulated in the MD.
Clusterization of inlets is performed in following steps:
Initial clusterization: All inlets are submitted to selected clusterization method and depending on the method and settings, some of the inlets might not be arranged to any cluster and are considered as outliers.
[Optional] Outliers detection: Arrangement of inlets to clusters is sometimes far from optimal. In this step, inlets that do not fit to cluster are detected and annotated as outliers. This step can be executed in two modes:
- Automatic mode: Inlet is considered to be an outlier if its distance from the centroid is greater then mean distance + 4 * standard deviation of all distances within the cluster.
- Defined threshold: Inlet is considered to be an outlier if its minimal distance from any other point in the cluster is greater then the threshold.
[Optional] Reclusterization of outliers: It may happen that the outliers form actually clusters but it was not recognized in initial clusterization. In this step clusterization is executed for outliers only and found clusters are appended to the clusters identified in the first step. Rest of the inlets are marked as outliers.
Potentially recursive clusterization¶
Both Initial clusterization and Reclustarization can be run in a recursive manner. If in the appropriate sections defining clusterization methods option recursive_clusterization is used appropriate method is run for each cluster separately. Clusters of specific size can be excluded from recursive clusterization (option recursive_threshold). It is also possible to limit maximal number of recursive levels - option max_level.
For additional information see clusterization sections options.
Available methods¶
Aqua-Duct implements several clustering methods. The recommended method is barber method which bases on Auto Barber procedure. Rest of the methods are implemented with sklearn.cluster module:
aquaduct.geom.cluster.BarberCluster- default for Initial clusterization. It gives excellent results. For more information see barber clusterization method description.MeanShift- see also original publication Mean shift: a robust approach toward feature space analysis (doi:10.1109/34.1000236).DBSCAN- default for Reclusterization of outliers, see also original publication A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with NoiseAffinityPropagation- see also original publication Clustering by Passing Messages Between Data Points (doi:10.1126/science.1136800)KMeans- see also k-means++: The advantages of careful seeding, Arthur, David, and Sergei Vassilvitskii in Proceedings of the eighteenth annual ACM-SIAM symposium on Discrete algorithms, Society for Industrial and Applied Mathematics (2007), pages 1027-1035.Birch- see also Tian Zhang, Raghu Ramakrishnan, Maron Livny BIRCH: An efficient data clustering method for large databases and Roberto Perdisci JBirch - Java implementation of BIRCH clustering algorithm.
For additional information see clusterization sections options.
Master paths¶
At the end of clusterization stage it is possible to run procedure for master path generation. First, separate paths are grouped according to clusters. Paths that begin and end in particular clusters are grouped together. Next, for each group a master path (i.e., average path) is generated in following steps:
First, length of master path is determined. Lengths of each parts (incoming, object, outgoing) for each separate paths are normalized with bias towards longest paths. These normalized lengths are then used for as weights in averaging not normalized lengths. Values for all parts are summed and resulting value is the desired length of master path.
All separate paths are divided into chunks. Number of chunks is equal to the desired length of master path calculated in the previous step. Lengths of separate paths can be quite diverse, therefore, for different paths chunks are of different lengths.
For each chunk averaging procedure is run:
- Coordinates for all separate paths for given chunk are collected.
- Normalized lengths with bias toward longest paths for all separate paths for given chunk are collected.
- New coordinates are calculated as weighted average of collected coordinates. As weights collected normalized lengths are used.
- In addition width of chunk is calculated as a mean value of collected coordinates mutual distances.
- Type of chunk is calculated as probability (frequency) of being in the scope.
Results for all chunks are collected, types probability are changed to types. All data is then used to create Master Path. If this fails no path is created.
More technical details on master path generation can be found in aquaduct.geom.master.CTypeSpathsCollection.get_master_path() method documentation.
Analysis¶
Fifth stage of Valve calculations analyses results calculated in stages 1 to 4. Results of the analysis are displayed on the screen or can be saved to text file and comprise of following parts:
- Tile and data stamp.
- [Optional] Dump of configuration options.
- Basic information on traceable residues and separate paths.
- Number of traceable residues.
- Number of separate paths.
- Basic information on inlets.
- Number of inlets.
- Number of clusters.
- Are outliers detected.
- Summary of inlets clusters. Table with 5 columns:
- Nr: Row number, starting from 0.
- Cluster: ID of the cluster. Outliers have 0.
- Size: Size of the cluster.
- INCOMING: Number of inlets corresponding to separate paths that enter the scope.
- OUTGOING: Number of inlets corresponding to separate paths that leave the scope.
- Summary of separate paths clusters types. Table with 9 columns.
- Nr: Row number, starting from 0.
- CType: Separate path Cluster Type.
- Size: Number of separate paths belonging to Cluster type.
- Tot: Average total length of the path.
- TotStd: Standard deviation of length Tot.
- Inp: Average length of incoming part of the path. If no incoming part is available it is NaN (not a number).
- InpStd: Standard deviation of length Inp.
- Obj: Average length of object part of the path. If no incoming part is available it is NaN.
- ObjStd: Standard deviation of length Inp.
- Out: Average length of outgoing part of the path. If no incoming part is available it is NaN.
- OutStd: Standard deviation of length Inp.
- List of separate paths and their properties. Table with 17 columns.
- Nr: - Row number, starting from 0.
- ID: - Separate path ID.
- RES: - Residue name.
- BeginF: Number of frame in which the path begins.
- InpF: Number of frame in which path begins Incoming part.
- ObjF: Number of frame in which path begins Object part.
- OutF: Number of frame in which path begins Outgoing part.
- EndF: Number of frame in which the path ends.
- TotL: Total length of path.
- InpL: Length of Incoming part. If no incoming part NaN is given.
- ObjL: Length of Object part.
- OutL: Length of Outgoing part. If no outgoing part NaN is given.
- TotS: Average step of full path.
- TotStdS: Standard deviation of TotS.
- InpS: Average step of Incoming part. If no incoming part NaN is given.
- InpStdS: Standard deviation of InpS.
- ObjS: Average step of Object part.
- ObjStdS: Standard deviation of ObjS.
- OutS: Average step of Outgoing part. If no outgoing part NaN is given.
- OutStdS: Standard deviation of OutS.
- CType: Cluster type of separate path.
Separate path ID¶
Separate Path IDs are composed of two numbers separated by colon. First number is the residue number. Second number is consecutive number of the separate path made by the residue. Numeration starts with 0.
Cluster Type of separate path¶
Each separate path has two ends: beginning and end. Both of them either belong to one of the clusters of inlets, or are among outliers, or are inside the scope. If an end belongs to one of the clusters (including outliers) it has ID of the cluster. If it is inside the scope it has special ID of N. Cluster type is an ID composed of IDs of both ends of separate path separated by colon charter.
Visualization¶
Sixth stage of Valve calculations visualizes results calculated in stages 1 to 4. Visualization is done with PyMOL. Valve automatically starts PyMOL and loads visualizations in to it. Molecule is loaded as PDB file. Other objects like Inlets clusters or paths are loaded as CGO objects.
Following is a list of objects created in PyMOL (all of them are optional). PyMOL object names given in bold text or short explanation is given.
- Selected frame of the simulated system. Object name: molecule.
- Inlets clusters, each cluster is a separate object. Object name: cluster_ followed by cluster annotation: otliers are annotated as Out; regular clusters by ID.
- List of cluster types, raw paths. Each cluster type is a separate object. Object name composed of cluster type (colon replaced by underline) plus _raw.
- List of cluster types, smooth paths. Each cluster type is a separate object. Object name composed of cluster type (colon replaced by underline) plus _smooth.
- All raw paths. They can be displayed as one object or separated in to Incoming, Object and Outgoing part. Object name: all_raw, or all_raw_in, all_raw_obj, and all_raw_out.
- All raw paths inlets arrows. Object name: all_raw_paths_io.
- All smooth paths. They can be displayed as one object or separated in to Incoming, Object and Outgoing part. Object name: all_smooth, or all_smooth_in, all_smooth_obj, and all_smooth_out.
- All raw paths inlets arrows. Object name: all_raw_paths_io.
- Raw paths displayed as separate objects or as one object with several states. Object name: raw_paths_ plus number of path or raw_paths if displayed as one object.
- Smooth paths displayed as separate objects or as one object with several states. Object name: smooth_paths_ plus number of path or smooth_paths if displayed as one object.
- Raw paths arrows displayed as separate objects or as one object with several states. Object name: raw_paths_io_ plus number of path or raw_paths_io if displayed as one object.
- Smooth paths arrows displayed as separate objects or as one object with several states. Object name: smooth_paths_io_ plus number of path or smooth_paths_io if displayed as one object.
Color schemes¶
Inlets clusters are colored automatically. Outliers are gray.
Incoming parts of paths are red, Outgoing parts are blue. Object parts in case of smooth paths are green and in case of raw paths are green if residue is precisely in the object area or yellow if is leaved object area but it is not in the Outgoing part yet. Passing paths are displayed in grey.
Arrows are colored in accordance to the colors of paths.
