Running pybool¶
We illustrate how pybool works using an example from a paper by Nakajima et al. The code that configures pybool to run this example is included in the pybool distribution. Later on we will show how to write your own code to configure pybool to run on your biological system of interest but for the time being let’s start with a system that has already been configured.
Nakajima et al. are interested in the regulatory network that controls neurogenesis in Drosophila. The system consists of six genes, hb, Kr, pdm, cas, svp and X. They have loss-of-function and over-expression time-course data for hb, Kr, pdm and cas. svp and X are regarded as external inputs to the system. In total this makes nine conditions: a wild-type condition and loss-of-function conditions and over-expression conditions for each of hb, Kr, pdm and cas. For each of the nine conditions Nakajima et al. have a set of expression constraints detailing the order in which genes are expressed. All of this information is encoded in a python module that is used to configure pybool: pybool.examples.tutorial.
To run this example execute the following command:
pybool-find-consistent --plot 5 pybool.examples.tutorial
This will generate some output to the screen and also create a directory called pyboolOutput which will contain several images and a log file.
Input: Restrictions¶
In pybool expert knowledge is encoded as restrictions on the permitted regulatory relationships. These restrictions are part of the configuration used as input to pybool. pybool will create graphical representations of which regulatory relationships are permitted in a file called net-restrictions.png.
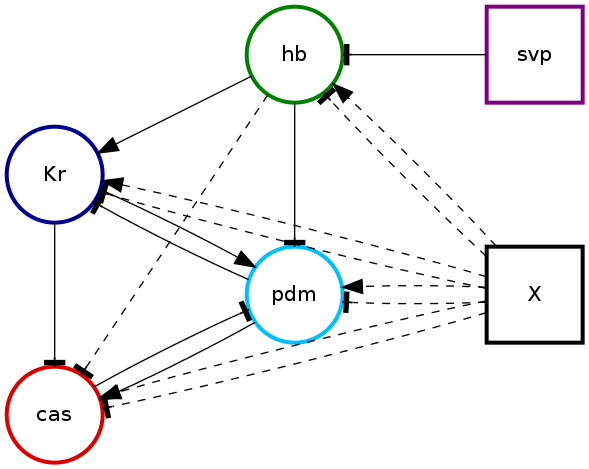
The first thing to notice is that every gene is configured to be represented by a specific color. This will be consistent across all of pybool’s output. Secondly, external inputs are represented as square nodes in the graph. Genes that can be regulated are represented as circles. The possible activatory or repressive regulatory relationships are shown as directed edges in the graph. Activatory relationships are represented by an arrowed edge, Repressive relationships are represented by edges that end in a bar. If the absence of regulation is allowed then the edges are dashed rather than solid.
We can see the expert knowledge encoded in the configuration. For example:
- hb represses pdm
- svp represses hb
- pdm activates cas
- cas represses pdm
- hb can repress cas or have no effect
- X can repress or activate or have no effect on pdm
Output: Consistent networks¶
There will also be a representation of the consistent networks that pybool found in the file net-consistent.png.
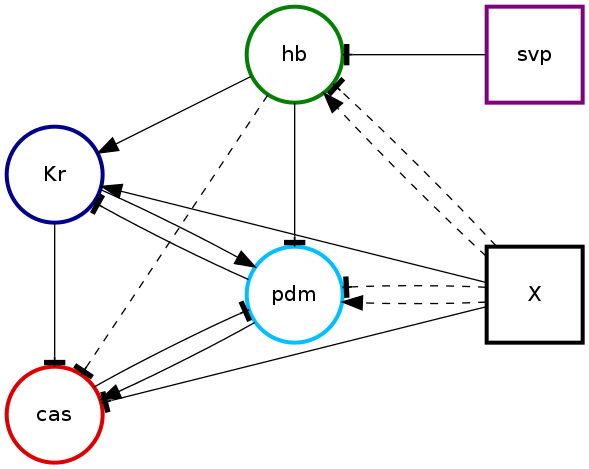
This image will show which edges were present in the set of consistent networks. The consistent networks are those which produce expression patterns (a.k.a. realisations) that are consistent with the temporal expression constraints specified in the input configuration.
Output: Example realisations¶
As we used the --plot 5 argument to pybool-find-consistent there will also be images representing five of the consistent networks and their realisations. Here is the second of these networks
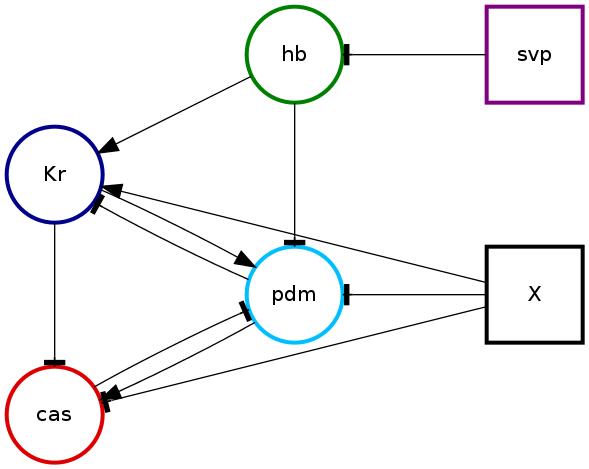
and its realisation
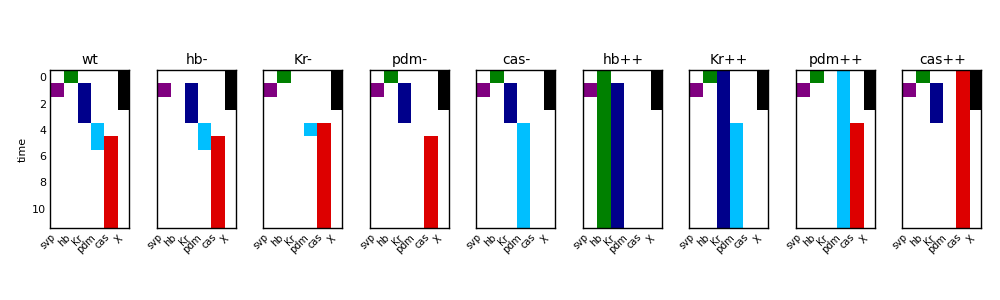
The realisation of each condition is represented separately. The genes are colour coded as in the network diagrams and the y-axis represents time from top to bottom.
Output: text¶
pybool also outputs information to the screen and a log file pybool-find-consistent.log. Here you can find extra information such as how many networks were tested for consistency, how many were consistent, which parameters for external inputs were in consistent networks, which constitutive expression levels were in consistent networks.
Added FileHandler logging to pyboolOutput/pybool-find-consistent.log.
2013-03-02 13:10:00,700:INFO:Options:
2013-03-02 13:10:00,700:INFO: parallel: None * Use ipython controller and engines to run in parallel.
2013-03-02 13:10:00,700:INFO: output_dir: pyboolOutput * Directory to store output in.
2013-03-02 13:10:00,700:INFO: max_to_try: None * Maximum number of networks to try. 0 for all.
2013-03-02 13:10:00,700:INFO: plot_networks: 5 * Number of networks to plot, with their realisations. -1 for all, 0 for none.
2013-03-02 13:10:00,700:INFO: black_and_white: None * Plot graphs in black and white.
2013-03-02 13:10:00,700:INFO: use_latex: None * Plot publication quality graphs using dot2tex and the LaTeX tikz package.
2013-03-02 13:10:00,700:INFO: formats: ['png'] * Plot graphs in given format (e.g. png, eps, svg, pdf). You can have more than one -F option on the command line.
2013-03-02 13:10:00,700:INFO:Importing constraints from python module pybool.examples.tutorial
2013-03-02 13:10:00,701:INFO:Have 6 genes called: svp,hb,Kr,pdm,cas,X
2013-03-02 13:10:00,701:INFO:The possible regulatory relationships are:
[[ svp hb Kr pdm cas X ]
[ svp 0 0 0 0 0 0 ]
[ hb - 0 0 0 0 -/0/+ ]
[ Kr 0 + 0 - 0 -/0/+ ]
[ pdm 0 - + 0 - -/0/+ ]
[ cas 0 -/0 - + 0 -/0/+ ]
[ X 0 0 0 0 0 0 ]]
2013-03-02 13:10:00,701:INFO:Gene hb : initial state 1 : constitutive expression (0, 1)
2013-03-02 13:10:00,701:INFO:Gene Kr : initial state 0 : constitutive expression (0, 1)
2013-03-02 13:10:00,701:INFO:Gene pdm : initial state 0 : constitutive expression (0, 1)
2013-03-02 13:10:00,701:INFO:Gene cas : initial state 0 : constitutive expression (0, 1)
2013-03-02 13:10:00,701:INFO: svp is an external input with possible input parameters: None
2013-03-02 13:10:00,702:INFO: X is an external input with possible input parameters: 1,2,3,4,5,6,7,8,9,10,11
2013-03-02 13:10:00,702:INFO:The conditions to test are: wt, hb-, Kr-, pdm-, cas-, hb++, Kr++, pdm++, cas++
2013-03-02 13:10:00,733:INFO:Running constraints 1 engines id=-1
2013-03-02 13:10:00,734:INFO:Will generate 28512 different networks.
2013-03-02 13:10:03,825:INFO:Evaluated 28512 networks in 3.1 seconds. 9221.0/sec
2013-03-02 13:10:03,825:INFO:Total mismatches = 146040
2013-03-02 13:10:03,929:INFO:Found 28512 networks which match the default condition "0" (161 distinct Js).
2013-03-02 13:10:03,930:INFO:Found 149 consistent networks which match all conditions (17 distinct Js).
2013-03-02 13:10:03,930:INFO:Condition broken count: wt = 26873
2013-03-02 13:10:03,930:INFO:Condition broken count: hb- = 995
2013-03-02 13:10:03,930:INFO:Condition broken count: Kr- = 243
2013-03-02 13:10:03,930:INFO:Condition broken count: pdm- = 252
2013-03-02 13:10:03,930:INFO:Condition broken count: cas- = 0
2013-03-02 13:10:03,930:INFO:Condition broken count: hb++ = 0
2013-03-02 13:10:03,930:INFO:Condition broken count: Kr++ = 0
2013-03-02 13:10:03,930:INFO:Condition broken count: pdm++ = 0
2013-03-02 13:10:03,930:INFO:Condition broken count: cas++ = 0
2013-03-02 13:10:03,930:INFO:Pickling good networks to all-conditions-networks.pickle
2013-03-02 13:10:03,943:INFO:Plotting 5 network(s) and realisations.
2013-03-02 13:10:07,685:INFO:Consistent regulatory relationships in the networks are:
[[ svp hb Kr pdm cas X ]
[ svp 0 0 0 0 0 0 ]
[ hb - 0 0 0 0 -/0/+ ]
[ Kr 0 + 0 - 0 + ]
[ pdm 0 - + 0 - -/0/+ ]
[ cas 0 -/0 - + 0 - ]
[ X 0 0 0 0 0 0 ]]
2013-03-02 13:10:07,695:INFO:How often X regulates hb 0 : 46% + : 6% - : 46%
2013-03-02 13:10:07,696:INFO:How often X regulates pdm 0 : 26% + : 47% - : 26%
2013-03-02 13:10:07,696:INFO:How often hb regulates cas 0 : 48% - : 51%
2013-03-02 13:10:07,697:INFO:Consistent constitutive expression levels for svp are 0
2013-03-02 13:10:07,697:INFO:Consistent constitutive expression levels for hb are 0
2013-03-02 13:10:07,697:INFO:Consistent constitutive expression levels for Kr are 0
2013-03-02 13:10:07,697:INFO:Consistent constitutive expression levels for pdm are 0,1
2013-03-02 13:10:07,697:INFO:Consistent constitutive expression levels for cas are 1
2013-03-02 13:10:07,697:INFO:Consistent constitutive expression levels for X are 0
2013-03-02 13:10:07,697:INFO:Counts for constitutive expression levels for pdm are 0 : 21% 1 : 78%
2013-03-02 13:10:07,698:INFO:Consistent input parameters for the external input of svp are: None
2013-03-02 13:10:07,698:INFO:Consistent input parameters for the external input of X are: 1,2,3,4,5,6,7,8,9
2013-03-02 13:10:07,698:INFO:Counts for input parameters for the external input of X are 1 : 10% 2 : 14% 3 : 10% 4 : 10% 5 : 10% 6 : 10% 7 : 10% 8 : 10% 9 : 10%
2013-03-02 13:10:07,821:INFO:Mutual information between theta:3 and J:(5, 3) is 0.19
2013-03-02 13:10:07,822:INFO:Mutual information between input parameters:5 and J:(5, 1) is 0.09
2013-03-02 13:10:07,822:INFO:Mutual information between theta:3 and input parameters:5 is 0.03
2013-03-02 13:10:07,822:INFO:Mutual information between J:(1, 4) and J:(5, 1) is 0.00
2013-03-02 13:10:07,822:INFO:Mutual information between theta:0 and input parameters:5 is 0.00
2013-03-02 13:10:07,822:INFO:Mutual information between theta:1 and input parameters:5 is 0.00
2013-03-02 13:10:07,822:INFO:Mutual information between theta:2 and input parameters:5 is 0.00
2013-03-02 13:10:07,822:INFO:Mutual information between theta:4 and input parameters:5 is 0.00
2013-03-02 13:10:07,822:INFO:Mutual information between theta:5 and input parameters:5 is 0.00
2013-03-02 13:10:07,823:INFO:Mutual information between input parameters:0 and input parameters:5 is 0.00
2013-03-02 13:10:07,823:INFO:Mutual information between input parameters:1 and input parameters:5 is 0.00
2013-03-02 13:10:07,823:INFO:Mutual information between input parameters:2 and input parameters:5 is 0.00
2013-03-02 13:10:07,823:INFO:Mutual information between input parameters:3 and input parameters:5 is 0.00
2013-03-02 13:10:07,823:INFO:Mutual information between input parameters:4 and input parameters:5 is 0.00
2013-03-02 13:10:07,823:INFO:Mutual information between J:(0, 0) and J:(5, 3) is 0.00
2013-03-02 13:10:07,823:INFO:Mutual information between J:(0, 1) and J:(5, 3) is 0.00
2013-03-02 13:10:07,823:INFO:Mutual information between J:(0, 2) and J:(5, 3) is 0.00
2013-03-02 13:10:07,823:INFO:Mutual information between J:(0, 3) and J:(5, 3) is 0.00
2013-03-02 13:10:07,823:INFO:Mutual information between J:(0, 4) and J:(5, 3) is 0.00
2013-03-02 13:10:07,824:INFO:Mutual information between J:(0, 5) and J:(5, 3) is 0.00
2013-03-02 13:10:07,824:INFO:Mutual information between J:(1, 0) and J:(5, 3) is 0.00
2013-03-02 13:10:07,824:INFO:Mutual information between J:(1, 1) and J:(5, 3) is 0.00
2013-03-02 13:10:07,824:INFO:Mutual information between J:(1, 2) and J:(5, 3) is 0.00
2013-03-02 13:10:07,824:INFO:Mutual information between J:(1, 3) and J:(5, 3) is 0.00
2013-03-02 13:10:07,824:INFO:Mutual information between J:(1, 5) and J:(5, 3) is 0.00
2013-03-02 13:10:07,824:INFO:Mutual information between J:(2, 0) and J:(5, 3) is 0.00
2013-03-02 13:10:07,824:INFO:Mutual information between J:(2, 1) and J:(5, 3) is 0.00
2013-03-02 13:10:07,824:INFO:Mutual information between J:(2, 2) and J:(5, 3) is 0.00
2013-03-02 13:10:07,825:INFO:Mutual information between J:(2, 3) and J:(5, 3) is 0.00
2013-03-02 13:10:07,825:INFO:Mutual information between J:(2, 4) and J:(5, 3) is 0.00
2013-03-02 13:10:07,825:INFO:Mutual information between J:(2, 5) and J:(5, 3) is 0.00
2013-03-02 13:10:07,825:INFO:Mutual information between J:(3, 0) and J:(5, 3) is 0.00
2013-03-02 13:10:07,825:INFO:Mutual information between J:(3, 1) and J:(5, 3) is 0.00
2013-03-02 13:10:07,825:INFO:Mutual information between J:(3, 2) and J:(5, 3) is 0.00
2013-03-02 13:10:07,825:INFO:Mutual information between J:(3, 3) and J:(5, 3) is 0.00
2013-03-02 13:10:07,825:INFO:Mutual information between J:(3, 4) and J:(5, 3) is 0.00
2013-03-02 13:10:07,825:INFO:Mutual information between J:(3, 5) and J:(5, 3) is 0.00
2013-03-02 13:10:07,826:INFO:Mutual information between J:(4, 0) and J:(5, 3) is 0.00
2013-03-02 13:10:07,826:INFO:Mutual information between J:(4, 1) and J:(5, 3) is 0.00
2013-03-02 13:10:07,826:INFO:Mutual information between J:(4, 2) and J:(5, 3) is 0.00
2013-03-02 13:10:07,826:INFO:Mutual information between J:(4, 3) and J:(5, 3) is 0.00
2013-03-02 13:10:07,826:INFO:Mutual information between J:(4, 4) and J:(5, 3) is 0.00
2013-03-02 13:10:07,826:INFO:Mutual information between J:(4, 5) and J:(5, 3) is 0.00
2013-03-02 13:10:07,826:INFO:Mutual information between J:(5, 0) and J:(5, 3) is 0.00
2013-03-02 13:10:07,826:INFO:Mutual information between J:(5, 2) and J:(5, 3) is 0.00
2013-03-02 13:10:07,826:INFO:Mutual information between input parameters:5 and J:(5, 4) is 0.00
2013-03-02 13:10:07,827:INFO:Mutual information between input parameters:5 and J:(5, 5) is 0.00
Parallelism¶
Finding consistent networks is easily parallelisable and pybool takes advantage of this using the IPython parallel framework. Full instructions for setting up this framework can be found on the IPython website but we describe a simple set up here. Make sure IPython is installed and start a cluster of four local engines:
ipcluster start -n 4
Now simply run pybool-find-consistent with the -p command line option:
pybool-find-consistent -p --plot 5 pybool.examples.tutorial
Here we have just used four engines on our local machine but IPython makes it easy to distribute the work across many engines on many machines. See their website for how to set this up.