
What is CrossMap ?¶
- CrossMap is a program for convenient conversion of genome coordinates (or annotation files) between different assemblies (such as Human hg18 (NCBI36) <> hg19 (GRCh37), Mouse mm9 (MGSCv37) <> mm10 (GRCm38)).
- It supports most commonly used file formats including SAM/BAM, Wiggle/BigWig, BED, GFF/GTF, VCF.
- CrossMap is designed to liftover genome coordinates between assemblies. It’s not a program for aligning sequences to reference genome.
- We do not recommend using CrossMap to convert genome coordinates between species.
Why CrossMap ?¶
Full genome sequencing, especially mammalian (eg. human) genomes, requires extensive, continuous efforts. Therefore reference genome assemblies are subject to change and refinement from time to time. Generally, researchers need to convert results that have been analyzed according to old assemblies to newer versions or vice versa, to facilitate meta-analysis, direct comparison as well as data integration and visualization.
Several useful conversion tools have been developed:
- UCSC liftover tool only supports BED input.
- NCBI remap support BED, GFF, GTF, VCF, etc
- Galaxy (Based on UCSC liftover tool) supports BED, GFF, GTF input.
- Ensembl assembly converter supports BED, GFF, GTF, PSL input, but output is GFF only. (Update: The original “assembly converter” has been retired. Starting from 2015, Ensembl uses CrossMap to perform genome coordinate conversion.)
- pyliftover “only does conversion of point coordinates, that is, unlike liftOver, it does not convert ranges, nor does it provide any special facilities to work with BED files”.
But none have the functionality to convert files in BAM/SAM or BigWig format. This is a significant gap in computational genomics tools, since these formats are the ones most widely used for representing high-throughput sequencing data such as RNA-seq, ChIP-seq, DNA-seq, etc.
Who is using CrossMap ?¶
How CrossMap works?¶
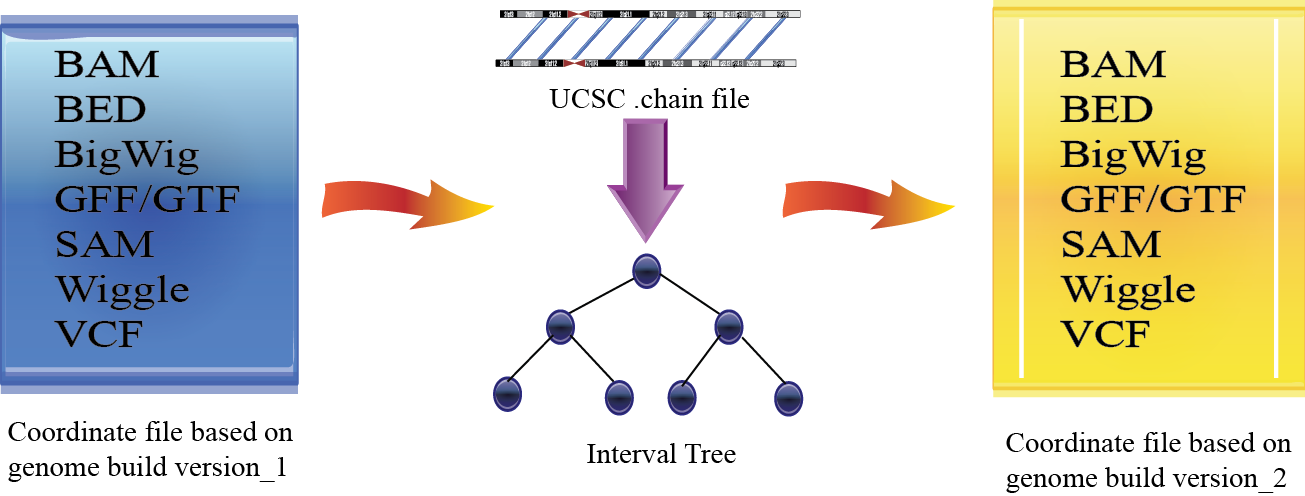
Algorithm¶
CrossMap first determines the correspondence between genome assemblies from UCSC chain file (chain file describes the pair-wise alignments between two genomes). Genome intervals will be stored in interval tree data structure, which allows one to efficiently find all intervals that overlap with any given interval or point. Then CrossMap remaps each entry in BAM/SAM, BED, GFF/GTF, VCF file to the target assembly by querying the interval tree. Exon/intron structure in BED file; spliced alignments, paired alignments, insert size, header section, SAM flags in BAM/SAM file; reference alleles, indels in VCF file will be processed properly.
For Wiggle/BigWig format files, line-by-line computation will be very slow. To increase speed, CrossMap groups consecutive coordinates with the same coverage score into bins (i.e. genomic regions), then remaps those regions one-by-one to the target assembly by querying the interval tree. In other words, Wiggle/BigWig files will be converted into bedGraph format internally, which will be converted into BigWig format (if UCSC’s ‘wigToBigWig‘ executable exists and is callable).
Time complexity¶
Assume there are N lines in the chain file. CrossMap loads the chain file first and process the query file line by line. Thus the space complexity is O(N). For each query region (s,t), it takes O(logN) time to locate which chain(s) overlap with s and t. Then it takes O(logN) time to search the sorted ungapped alignments in this chain that overlap with s and t and calculate the converted values for s and t in the target assembly. So in total it takes O(logN) time to convert one query. The time complexity is O(logN*M) to convert M queries.
In practical, the time CrossMap takes increases linearly to the size of input file.
Release history¶
- 05/09/17: Release version 0.2.6:
- In BAM file liftover: fixed bugs during BAM file sorting and indexing steps (works with pysam v0.11.1).
- In BAM file liftover: fixed bugs “the read group type is automatically and wrongly changed from Z to A” (https://github.com/pysam-developers/pysam/issues/113).
- 10/7/16: Release version 0.2.5:
- fixed bugs during single-end BAM file conversion.
- Add optional tags to the output BAM file. Details see: Convert BAM/SAM format files.
- 08/18/16: Release version 0.2.4:
- fixed bugs during BAM file conversion:
- When the strand of read changes, the seq filed is reverse complemented and the quality field is reversed.
- In the output VCF file, if the reference allele field is empty:
- Use CrossMap v0.2.4. Update pysam to the latest version. And make sure chromosome IDs in the reference genome file are in the form of “chr1”, “chr2”, ..., “chrX”,”chrY” (but not “1”, “2”, ..., “X”,”Y”, in this case, pysam cannot index your reference genome file for some unknown reasons.).
- to upgrade, run: pip install CrossMap –upgrade
- 04/13/16: Release version 0.2.3:
- Same as v0.2.2.
- Two dependency packages bx-python and pysam do not shipped with CrossMap starting from v0.2.3 .
- Users could install CrossMap using pip: pip install CrossMap. Note: bx-python and pysam will be installed automatically if they haven’t been installed before.
- 11/10/15: Release version 0.2.2: Generate *.unmap files (regions that cannot be unambiguously converted) when converting BED, GTF, GFF files. This version also supports genePred (bed12+8) format. (Thanks for Andrew Yates from EMBL-EBI)
- 08/26/15: Release version 0.2.1: Very minor change, same as 0.2.
- 08/11/15: Release version 0.2: Fixed the bug that CrossMap will not convert wiggle format files due to name collision with bx python.
- 07/27/15: Release version 0.1.9. For VCF file conversion in v0.1.9:
- CrossMap uses the indexed reference genome (target assembly) sequences rather than load the entire file into memory. Users could index their reference genome file using samtools faidx before running CrossMap, otherwise CrossMap will index it automatically the first time you run it.
- In the output VCF file, whether the chromosome IDs contain “chr” or not depends on the input format.
- 05/15/15: Release version 0.1.8: Fixed the bug that CrossMap will output invalid VCF file when the input VCF file contains a INFO field with whitespace.
- 05/04/15: Release version 0.1.7: Address the problem that CrossMap does not convert strand in inversions when input file is BED6 or BED12 format.
- 11/06/14: Release version 0.1.6: Fixed “negative coordinates” bug.
- 08/05/14: Release version 0.1.5: Support compressed (*.gz, *.Z, *.z, *.bz, *.bz2, *.bzip2) wiggle file as input.
- 05/19/14: add chain files for hg38->hg19, hg19->hg38, hg18->hg38, hg19->GRCh37, GRCh37->hg19. In CrossMap v0.1.4, conversion results of BAM/SAM files can be directed to STDOUT to support piping.
- 12/12/13: CrossMap was accepted by Bioinformatics
- 10/23/13: CrossMap (0.1.3) was released
Installation¶
Use pip to install CrossMap¶
pip install CrossMap
Use pip to upgrade CrossMap¶
pip install CrossMap --upgrade
Install CrossMap from source code¶
- Source code CrossMap
- Test datsets
Prerequisite:
$ tar zxf CrossMap-VERSION.tar.gz
$ cd CrossMap-VERSION
# install CrossMap to default location. In Linux/Unix, this location is like:
# /home/user/lib/python2.7/site-packages/
$ python setup.py install
# or you can install CrossMap to a specified location:
$ python setup.py install --root=/home/user/CrossMap
# setup PYTHONPATH. Skip this step if CrossMap was installed to default location.
$ export PYTHONPATH=/home/user/CrossMap/usr/local/lib/python2.7/site-packages:$PYTHONPATH.
# Skip this step if CrossMap was installed to default location.
$ export PATH=/home/user/CrossMap/usr/local/bin:$PATH
NOTE:
- Mac users need to download and install Xcode command line tools.
Input and Output¶
CrossMap basically needs 2 input files. chain format file describing genom-wide pairwise alignments between assemblies and the file containing genome coordinates that you want to convert to different assembly. If input file is in VCF format, a reference genome sequence file(in FASTA format) is needed.
Chain file¶
Example of chain file:
chain 4900 chrY 58368225 + 25985403 25985638 chr5 151006098 - 43257292 43257528 1
9 1 0
10 0 5
61 4 0
16 0 4
42 3 0
16 0 8
14 1 0
3 7 0
48
chain 4900 chrY 58368225 + 25985406 25985566 chr5 151006098 - 43549808 43549970 2
16 0 2
60 4 0
10 0 4
70
UCSC built chain files (Human, Homo sapiens)
- hg38ToHg19.over.chain.gz (Chain file needed to convert hg38 to hg19)
- hg19ToHg38.over.chain.gz (Chain file needed to convert hg19 to hg38)
- hg18ToHg38.over.chain.gz (Chain file needed to convert hg18 to hg38)
- hg19ToHg18.over.chain.gz (Chain file needed to convert hg19 to hg18)
- hg19ToHg17.over.chain.gz (Chain file needed to convert hg19 to hg17)
- hg18ToHg19.over.chain.gz (Chain file needed to convert hg18 to hg19)
- hg18ToHg17.over.chain.gz (Chain file needed to convert hg18 to hg17)
- hg17ToHg19.over.chain.gz (Chain file needed to convert hg17 to hg19)
- hg17ToHg18.over.chain.gz (Chain file needed to convert hg17 to hg18)
- GRCh37ToHg19.over.chain.gz (Chain file needed to convert GRCh37 to hg19)
- hg19ToGRCh37.over.chain.gz (Chain file needed to convert hg19 to GRCh37)
UCSC built chain files (Mouse, Mus musculus)
- mm10ToMm9.over.chain.gz (Chain file needed to convert mm10 to mm9)
- mm9ToMm10.over.chain.gz (Chain file needed to convert mm9 to mm10)
- mm9ToMm8.over.chain.gz (Chain file needed to convert mm9 to mm8)
UCSC Chain file of other species can be downloaded from: http://hgdownload.soe.ucsc.edu/downloads.html
Ensembl built chain files (Human, Homo sapiens)
- NCBI34 <=> GRCh38
- NCBI35 <=> GRCh38
- NCBI36 <=> GRCh38
- GRCh37 <=> GRCh38
- NCBI34 <=> GRCh37
- NCBI35 <=> GRCh37
- NCBI36 <=> GRCh37
Ensembl built chain files (Mouse, Mus musculus)
- NCBIM37_to_GRCm38.chain.gz
- GRCm38_to_NCBIM36.chain.gz
- GRCm38_to_NCBIM37.chain.gz
- NCBIM36_to_GRCm38.chain.gz
Ensembl Chain file of other species can be downloaded from: ftp://ftp.ensembl.org/pub/assembly_mapping/
User Input file¶
- BAM or SAM format.
- BED or BED-like format. BED file must has at least 3 columns (‘chrom’, ‘start’, ‘end’).
- Wiggle format. “variableStep”, “fixedStep” and “bedGraph” wiggle line are supported.
- BigWig format.
- GFF or GTF format.
- VCF format.
NOTE: When converting bedGraph file, Treat it as Wiggle format rather than BED format.
Output file¶
Format of Output files depends on the input format
| Input_format | Output_format |
|---|---|
| BED | BED (Genome coordinates will be updated to the target assembly) |
| BAM | BAM (Genome coordinates, header section, all SAM flags, insert size will be updated accordingly) |
| SAM | SAM (Genome coordinates, header section, all SAM flags, insert size will be updated accordingly) |
| Wiggle | bedGraph (if wigToBigWig executable does not exist) |
| Wiggle | BigWig (if wigToBigWig executable exists) |
| BigWig | bedGraph (if wigToBigWig executable does not exist) |
| BigWig | BigWig (if wigToBigWig executable exists) |
| GFF | GFF (Genome coordinates will be updated to the target assembly) |
| GTF | GTF (Genome coordinates will be updated to the target assembly) |
| VCF | VCF (Genome coordinates and reference alleles will be updated to the target assembly) |
Usage¶
Run CrossMap.py without any arguments will print help message:
# run CrossMap without argument
$ python CrossMap.py
Screen output:
Program: CrossMap (v0.1.1)
Description:
CrossMap is a program for convenient conversion of genome coordinates
and genomeannotation files between assemblies (eg. lift from human
hg18 to hg19 or vice versa).It support file in BAM, SAM, BED, Wiggle,
BigWig, GFF, GTF, VCF, etc.
Usage: CrossMap.py <command> [options]
bam convert alignment file in BAM or SAM format.
bed convert genome cooridnate or annotation file in BED or BED-like format.
bigwig convert genome coordinate file in BigWig format.
gff convert genome cooridnate or annotation file in GFF or GTF format.
vcf convert genome coordinate file in VCF format.
wig convert genome coordinate file in Wiggle, or bedGraph format.
Run CrossMap.py with command keyword will print help message for that command:
$ python CrossMap.py bed
Screen output:
Usage:
CrossMap.py bed input_chain_file input_bed_file [output_file]
Description:
"input_chain_file" and "input_bed_file" can be regular or compressed
(*.gz, *.Z, *.z, *.bz, *.bz2, *.bzip2) file, local file or URL
(http://, https://, ftp://) pointing to remote file. BED file must
have at least 3 columns (chrom, start, end) and no more than 12
columns. If no "output_file" was specified, output will be directed
to screen (console). BED format:
http://genome.ucsc.edu/FAQ/FAQformat.html#format1
Example:
CrossMapy.py bed hg18ToHg19.over.chain.gz test.hg18.bed test.hg19.bed
# write output to "test.hg19.bed"
Example:
CrossMapy.py bed hg18ToHg19.over.chain.gz test.hg18.bed
# write output to screen
Convert BED format files¶
A BED (Browser Extensible Data) file is a tab-delimited text file describing genome regions or gene annotations. It is the standard file format used by UCSC. It consists of one line per feature, each containing 3-12 columns. CrossMap converts BED files with less than 12 columns to a different assembly by updating the chromosome and genome coordinates only; all other columns remain unchanged. Regions from old assembly mapping to multiple locations to the new assembly will be split. For 12-columns BED files, all columns will be updated accordingly except the 4th column (name of bed line), 5th column (score value) and 9th column (RGB value describing the display color). 12-column BED files usually define multiple blocks (eg. exon); if any of the exons fails to map to a new assembly, the whole BED line is skipped.
The input BED file can be plain text file, compressed file with extension of .gz, .Z, .z, .bz, .bz2 and .bzip2, or even a URL pointing to accessible remote files (http://, https:// and ftp://). Compressed remote files are not supported. The output is a BED format file with exact the same number of columns as the original one.
Standard BED format has 12 columns, but CrossMap also supports BED-like formats:
- BED3: The first 3 columns (“chrom”, “start”, “end”) of BED format file.
- BED6: The first 6 columns (“chrom”, “start”, “end”, “name”, “score”, “strand”) of BED format file.
- Other: Format has at least 3 columns (“chrom”, “start”, “end”) and no more than 12 columns. All other columns are arbitrary.
NOTE:
- For BED-like formats mentioned above, CrossMap only updates “chrom (1st column)”, “start (2nd column) ”, “end (3rd column) ” and “strand” (if any). All other columns will keep AS-IS.
- Lines starting with ‘#’, ‘browser’, ‘track’ will be skipped.
- Lines will less than 3 columns will be skipped.
- 2nd-column and 3-column must be integer, otherwise skipped.
- “+” strand is assumed if no strand information was found.
- For standard BED format (12 columns). If any of the defined exon blocks cannot be uniquely mapped to target assembly, the whole entry will be skipped.
- “input_chain_file” and “input_bed_file” can be regular or compressed (.gz, .Z, .z, .bz, .bz2, .bzip2) file, local file or URL (http://, https://, ftp://) pointing to remote file.
- If output_file was not specified, results will be printed to screen (console). In this case, the original bed entries (include items failed to convert) were also printed out.
- If input region cannot be consecutively mapped target assembly, it will be split.
- *.unmap file contains regions that cannot be unambiguously converted.
Example (run CrossMap with no output_file specified):
$ python CrossMap.py bed hg18ToHg19.over.chain.gz test.hg18.bed3
Conversion results were printed to screen directly (column1-3 are hg18 based, column5-7 are hg19 based):
chr1 142614848 142617697 -> chr1 143903503 143906352
chr1 142617697 142623312 -> chr1 143906355 143911970
chr1 142623313 142623350 -> chr1 143911971 143912008
chr1 142623351 142626523 -> chr1 143912009 143915181
chr1 142633862 142633883 -> chr1 143922520 143922541
chr1 142633884 142636152 -> chr1 143922542 143924810
chr1 142636152 142636326 -> chr1 143924813 143924987
chr1 142636339 142636391 -> chr1 143925000 143925052
chr1 142636392 142637362 -> chr1 143925052 143926022
chr1 142637373 142639738 -> chr1 143926033 143928398
chr1 142639739 142639760 -> chr1 143928399 143928420
chr1 142639761 142640145 -> chr1 143928421 143928805
chr1 142640153 142641149 -> chr1 143928813 143929809
Example (run CrossMap with output_file (test.hg19.bed3) specified):
$ python CrossMap.py bed hg18ToHg19.over.chain.gz test.hg18.bed3 test.hg19.bed3
$ cat test.hg19.bed3
chr1 143903503 143906352
chr1 143906355 143911970
chr1 143911971 143912008
chr1 143912009 143915181
chr1 143922520 143922541
chr1 143922542 143924810
chr1 143924813 143924987
chr1 143925000 143925052
chr1 143925052 143926022
chr1 143926033 143928398
chr1 143928399 143928420
chr1 143928421 143928805
chr1 143928813 143929809
Example (one input region was split because it cannot be consecutively mapped target assembly):
$ python CrossMap.py bed hg18ToHg19.over.chain.gz test.hg18.bed3
chr10 81346644 81349952 + -> chr10 81356692 81360000 +
chr10 81349952 81364937 + -> chr10 81360000 81374985 +
chr10 81364952 81365854 + -> chr10 81375000 81375902 +
chr10 81365875 81369946 + -> chr10 81375929 81380000 +
chr10 81369946 81370453 + -> chr10 81380000 81380507 +
chr10 81370483 81371363 + -> chr10 81380539 81381419 +
chr10 81371363 81371365 + -> chr10 62961832 62961834 +
chr10 81371412 81371432 + (split.1:chr10:81371412:81371422:+) chr10 62961775 62961785 +
chr10 81371412 81371432 + (split.2:chr10:81371422:81371432:+) chrX 63278348 63278358 +
Convert BAM/SAM format files¶
SAM (Sequence Alignment Map) format is a generic format for storing sequencing alignments, and BAM is binary and compressed version of SAM (Li et al., 2009). Most high-throughput sequencing (HTS) alignments were in SAM/BAM format and many HTS analysis tools work with SAM/BAM format. CrossMap updates chromosomes, genome coordinates, header sections, and all SAM flags accordingly. The program version (of CrossMap) is inserted into the header section, along with the names of the original BAM file and the chain file. For pair-end sequencing, insert size is also recalculated. The input BAM file should be sorted and indexed properly using samTools (Li et al., 2009). Output format is determined from the input format and BAM output will be sorted and indexed automatically.
Typing command without any arguments will print help message:
$ python CrossMap.py bam
Screen output:
Usage: CrossMap.py bam input_chain_file input_bam_file output_file [options]
Note: If output_file == STDOUT or -, CrossMap will write BAM file to the screen
Options:
-m INSERT_SIZE, --mean=INSERT_SIZE
Average insert size of pair-end sequencing (bp).
[default=200.0]
-s INSERT_SIZE_STDEV, --stdev=INSERT_SIZE_STDEV
Stanadard deviation of insert size. [default=30.0]
-t INSERT_SIZE_FOLD, --times=INSERT_SIZE_FOLD
A mapped pair is considered as "proper pair" if both
ends mapped to different strand and the distance
between them is less then '-t' * stdev from the mean.
[default=3.0]
-a, --append-tags Add tag to each alignment.
Example (Convert BAM from hg19 to hg18):
$ CrossMap.py bam ../data/hg19ToHg18.over.chain.gz test.hg19.bam test.hg18 # do NOT add optional tags
Insert size = 200.000000
Insert size stdev = 30.000000
Number of stdev from the mean = 3.000000
Add tags to each alignment = False
@ 2016-10-07 15:29:06: Read chain_file: ../data/hg19ToHg18.over.chain.gz
@ 2016-10-07 15:29:07: Liftover BAM file: test.hg19.bam ==> test.hg18.bam
@ 2016-10-07 15:29:14: Done!
@ 2016-10-07 15:29:14: Sort "test.hg18.bam" ...
@ 2016-10-07 15:29:15: Index "test.hg18.sorted.bam" ...
Total alignments:99914
QC failed: 0
R1 unique, R2 unique (UU): 96094
R1 unique, R2 unmapp (UN): 3579
R1 unique, R2 multiple (UM): 0
R1 multiple, R2 multiple (MM): 0
R1 multiple, R2 unique (MU): 233
R1 multiple, R2 unmapped (MN): 8
R1 unmap, R2 unmap (NN): 0
R1 unmap, R2 unique (NU): 0
R1 unmap, R2 multiple (NM): 0
$ CrossMap.py bam -a ../data/hg19ToHg18.over.chain.gz test.hg19.bam test.hg18 # add optional tags using '-a'
Insert size = 200.000000
Insert size stdev = 30.000000
Number of stdev from the mean = 3.000000
Add tags to each alignment = True
@ 2016-10-07 15:29:06: Read chain_file: ../data/hg19ToHg18.over.chain.gz
@ 2016-10-07 15:29:07: Liftover BAM file: test.hg19.bam ==> test.hg18.bam
@ 2016-10-07 15:29:14: Done!
@ 2016-10-07 15:29:14: Sort "test.hg18.bam" ...
@ 2016-10-07 15:29:15: Index "test.hg18.sorted.bam" ...
Total alignments:99914
QC failed: 0
R1 unique, R2 unique (UU): 96094
R1 unique, R2 unmapp (UN): 3579
R1 unique, R2 multiple (UM): 0
R1 multiple, R2 multiple (MM): 0
R1 multiple, R2 unique (MU): 233
R1 multiple, R2 unmapped (MN): 8
R1 unmap, R2 unmap (NN): 0
R1 unmap, R2 unique (NU): 0
R1 unmap, R2 multiple (NM): 0
# BAM/SAM header sections was updated:
$ samtools view -H test.hg19.bam
@SQ SN:chr1 LN:249250621
@SQ SN:chr2 LN:243199373
@SQ SN:chr3 LN:198022430
@SQ SN:chr4 LN:191154276
@SQ SN:chr5 LN:180915260
@SQ SN:chr6 LN:171115067
@SQ SN:chr7 LN:159138663
@SQ SN:chr8 LN:146364022
@SQ SN:chr9 LN:141213431
@SQ SN:chr10 LN:135534747
@SQ SN:chr11 LN:135006516
@SQ SN:chr12 LN:133851895
@SQ SN:chr13 LN:115169878
@SQ SN:chr14 LN:107349540
@SQ SN:chr15 LN:102531392
@SQ SN:chr16 LN:90354753
@SQ SN:chr17 LN:81195210
@SQ SN:chr18 LN:78077248
@SQ SN:chr19 LN:59128983
@SQ SN:chr20 LN:63025520
@SQ SN:chr21 LN:48129895
@SQ SN:chr22 LN:51304566
@SQ SN:chrX LN:155270560
@SQ SN:chrY LN:59373566
@SQ SN:chrM LN:16571
@RG ID:Sample_618545BE SM:Sample_618545BE LB:Sample_618545BE PL:Illumina
@PG ID:bwa PN:bwa VN:0.6.2-r126
$ samtools view -H test.hg18.bam
@HD VN:1.0 SO:coordinate
@SQ SN:chr1 LN:247249719
@SQ SN:chr10 LN:135374737
@SQ SN:chr11 LN:134452384
@SQ SN:chr11_random LN:215294
@SQ SN:chr12 LN:132349534
@SQ SN:chr13 LN:114142980
@SQ SN:chr13_random LN:186858
@SQ SN:chr14 LN:106368585
@SQ SN:chr15 LN:100338915
@SQ SN:chr15_random LN:784346
@SQ SN:chr16 LN:88827254
@SQ SN:chr17 LN:78774742
@SQ SN:chr17_random LN:2617613
@SQ SN:chr18 LN:76117153
@SQ SN:chr18_random LN:4262
@SQ SN:chr19 LN:63811651
@SQ SN:chr19_random LN:301858
@SQ SN:chr1_random LN:1663265
@SQ SN:chr2 LN:242951149
@SQ SN:chr20 LN:62435964
@SQ SN:chr21 LN:46944323
@SQ SN:chr21_random LN:1679693
@SQ SN:chr22 LN:49691432
@SQ SN:chr22_random LN:257318
@SQ SN:chr3 LN:199501827
@SQ SN:chr3_random LN:749256
@SQ SN:chr4 LN:191273063
@SQ SN:chr4_random LN:842648
@SQ SN:chr5 LN:180857866
@SQ SN:chr6 LN:170899992
@SQ SN:chr6_random LN:1875562
@SQ SN:chr7 LN:158821424
@SQ SN:chr7_random LN:549659
@SQ SN:chr8 LN:146274826
@SQ SN:chr8_random LN:943810
@SQ SN:chr9 LN:140273252
@SQ SN:chr9_random LN:1146434
@SQ SN:chrM LN:16571
@SQ SN:chrX LN:154913754
@SQ SN:chrX_random LN:1719168
@SQ SN:chrY LN:57772954
@RG ID:Sample_618545BE SM:Sample_618545BE LB:Sample_618545BE PL:Illumina
@PG PN:bwa ID:bwa VN:0.6.2-r126
@PG ID:CrossMap VN:0.1.3
@CO Liftover from original BAM/SAM file: test.hg19.bam
@CO Liftover is based on the chain file: ../test/hg19ToHg18.over.chain.gz
Optional tags:
- Q
- QC. QC failed.
- N
- Unmapped. Originally unmapped or originally mapped but failed to liftover to new assembly.
- M
- Multiple mapped. Alignment can be liftover to multiple places.
- U
- Unique mapped. Alignment can be liftover to only 1 place.
Tags for pair-end sequencing include:
- QF = QC failed
- NN = both read1 and read2 unmapped
- NU = read1 unmapped, read2 unique mapped
- NM = read1 unmapped, multiple mapped
- UN = read1 uniquely mapped, read2 unmap
- UU = both read1 and read2 uniquely mapped
- UM = read1 uniquely mapped, read2 multiple mapped
- MN = read1 multiple mapped, read2 unmapped
- MU = read1 multiple mapped, read2 unique mapped
- MM = both read1 and read2 multiple mapped
Tags for single-end sequencing include:
- QF = QC failed
- SN = unmaped
- SM = multiple mapped
- SU = uniquely mapped
NOTE:
- Input is BAM or SAM format file. Output format depends on input format. (i.e BAM -> BAM, SAM -> SAM)
- Alignments that are failed to convert will be saved in “.unmap.bam” or ‘.unmap.sam‘.
- If output file is specified as “STDOUT”, output will be directed to the screen. (in this case, unmapped alignments will be saved to “input.unmap.bam” or “input.unmap.sam”)
- Header section will be updated to target assembly.
- Genome coordinates and all SAM flags in alignment section will be updated to target assembly.
- Optional fields in alignment section will not be updated in current version.
Convert Wiggle/BigWig format files¶
Wiggle (WIG) format is useful for displaying continuous data such as GC content and reads intensity of high-throughput sequencing data. BigWig is a self-indexed binary-format Wiggle file, and has the advantage of supporting random access. This means only regions that need to be displayed are retrieved by genome browser, and it dramatically reduces the time needed for data transferring (Kent et al., 2010). Input wiggle data can be in variableStep (for data with irregular intervals) or fixedStep (for data with regular intervals). Regardless of the input, the output will always in bedGraph format. bedGraph format is similar to wiggle format and can be converted into BigWig format using UCSC wigToBigWig tool. We export files in bedGraph because it is usually much smaller than file in wiggle format, and more importantly, CrossMap internally transforms wiggle into bedGraph to increase running speed.
If an input file is in BigWig format, the output is BigWig format if UCSC’s ‘wigToBigWig‘ executable can be found; otherwise, the output file will be in bedGraph format.
Typing command without any arguments will print help message:
$ python2.7 CrossMap.py wig
Screen output:
Usage:
CrossMap.py wig input_chain_file input_wig_file output_prefix
Description:
"input_chain_file" can be regular or compressed (*.gz, *.Z, *.z, *.bz, *.bz2,
*.bzip2) file, local file or URL (http://, https://, ftp://) pointing to remote
file. Both "variableStep" and "fixedStep" wiggle lines are supported. Wiggle
format: http://genome.ucsc.edu/goldenPath/help/wiggle.html
Example:
CrossMapy.py wig hg18ToHg19.over.chain.gz test.hg18.wig test.hg19
NOTE:
- To improve performance, this script calls GNU “sort” command internally. If “sort” command does not exist, CrossMap will exit.
Typing command without any arguments will print help message:
$ python2.7 CrossMap.py bigwig
Screen output:
Usage:
CrossMap.py bigwig input_chain_file input__bigwig_file output_prefix
Description:
"input_chain_file" can be regular or compressed (*.gz, *.Z, *.z, *.bz, *.bz2,
*.bzip2) file, local file or URL (http://, https://, ftp://) pointing to remote
file. Bigwig format: http://genome.ucsc.edu/goldenPath/help/bigWig.html
Example:
CrossMapy.py bigwig hg18ToHg19.over.chain.gz test.hg18.bw test.hg19
Example (Convert BigWig file from hg18 to hg19):
$ python CrossMap.py bigwig hg19ToHg18.over.chain.gz test.hg19.bw test.hg18
@ 2013-11-17 22:12:42: Read chain_file: ../data/hg19ToHg18.over.chain.gz
@ 2013-11-17 22:12:44: Liftover bigwig file: test.hg19.bw ==> test.hg18.bgr
@ 2013-11-17 22:15:38: Merging overlapped entries in bedGraph file ...
@ 2013-11-17 22:15:38: Sorting bedGraph file:test.hg18.bgr
@ 2013-11-17 22:15:39: Convert wiggle to bigwig ...
NOTE:
- To improve performance, this script calls GNU “sort” command internally. If “sort” command does not exist, CrossMap will exit.
- Output files: output_prefix.bw, output_prefix.bgr, output_prefix.sorted.bgr
Convert GFF/GTF format files¶
GFF (General Feature Format) is another plain text file used to describe gene structure. GTF (Gene Transfer Format) is a refined version of GTF. The first eight fields are the same as GFF. Plain text, compressed plain text, and URLs pointing to remote files are all supported. Only chromosome and genome coordinates are updated. The format of output is determined from the input.
Typing command without any arguments will print help message:
$ python2.7 CrossMap.py gff
Screen output:
Usage:
CrossMap.py gff input_chain_file input_gff_file output_file
Description:
"input_chain_file" can be regular or compressed (*.gz, *.Z, *.z, *.bz, *.bz2,
*.bzip2) file, local file or URL (http://, https://, ftp://) pointing to remote
file. input file must be in GFF or GTF format. GFF format:
http://genome.ucsc.edu/FAQ/FAQformat.html#format3 GTF format:
http://genome.ucsc.edu/FAQ/FAQformat.html#format4
Example:
CrossMap.py gff hg19ToHg18.over.chain.gz test.hg19.gtf test.hg18.gtf #write output to test.hg18.gtf
Example:
CrossMap.py gff hg19ToHg18.over.chain.gz test.hg19.gtf # write output to screen
Example (Convert GTF file from hg19 to hg18):
$ python CrossMap.py gff hg19ToHg18.over.chain.gz test.hg19.gtf test.hg18.gtf
@ 2013-11-17 20:44:47: Read chain_file: ../data/hg19ToHg18.over.chain.gz
$ head test.hg19.gtf
chr1 hg19_refGene CDS 48267145 48267291 0.000000 - 0 gene_id "NM_001194986"; transcript_id "NM_001194986";
chr1 hg19_refGene exon 66081691 66081907 0.000000 + . gene_id "NM_002303"; transcript_id "NM_002303";
chr1 hg19_refGene CDS 145334684 145334792 0.000000 + 2 gene_id "NM_001039703"; transcript_id "NM_001039703";
chr1 hg19_refGene exon 172017752 172017890 0.000000 + . gene_id "NM_001136127"; transcript_id "NM_001136127";
chr1 hg19_refGene CDS 206589249 206589333 0.000000 + 2 gene_id "NM_001170637"; transcript_id "NM_001170637";
chr1 hg19_refGene exon 210573812 210574006 0.000000 + . gene_id "NM_001170580"; transcript_id "NM_001170580";
chr1 hg19_refGene CDS 235850249 235850347 0.000000 - 0 gene_id "NM_000081"; transcript_id "NM_000081";
chr1 hg19_refGene CDS 235880012 235880078 0.000000 - 1 gene_id "NM_000081"; transcript_id "NM_000081";
chr1 hg19_refGene exon 3417741 3417872 0.000000 - . gene_id "NM_001409"; transcript_id "NM_001409";
chr1 hg19_refGene exon 10190773 10190871 0.000000 + . gene_id "NM_006048"; transcript_id "NM_006048";
$ head test.hg18.gtf
chr1 hg19_refGene CDS 48039732 48039878 0.000000 - 0 gene_id "NM_001194986"; transcript_id "NM_001194986";
chr1 hg19_refGene exon 65854279 65854495 0.000000 + . gene_id "NM_002303"; transcript_id "NM_002303";
chr1 hg19_refGene CDS 144046041 144046149 0.000000 + 2 gene_id "NM_001039703"; transcript_id "NM_001039703";
chr1 hg19_refGene exon 170284375 170284513 0.000000 + . gene_id "NM_001136127"; transcript_id "NM_001136127";
chr1 hg19_refGene CDS 204655872 204655956 0.000000 + 2 gene_id "NM_001170637"; transcript_id "NM_001170637";
chr1 hg19_refGene exon 208640435 208640629 0.000000 + . gene_id "NM_001170580"; transcript_id "NM_001170580";
chr1 hg19_refGene CDS 233916872 233916970 0.000000 - 0 gene_id "NM_000081"; transcript_id "NM_000081";
chr1 hg19_refGene CDS 233946635 233946701 0.000000 - 1 gene_id "NM_000081"; transcript_id "NM_000081";
chr1 hg19_refGene exon 3407601 3407732 0.000000 - . gene_id "NM_001409"; transcript_id "NM_001409";
chr1 hg19_refGene exon 10113360 10113458 0.000000 + . gene_id "NM_006048"; transcript_id "NM_006048";
NOTE:
- Each feature (exon, intron, UTR, etc) is processed separately and independently, and we do NOT check if features originally belonging to the same gene were converted into the same gene.
- If user want to liftover gene annotation files, use BED12 format.
- If no output file was specified, output will be printed to screen (console). In this case, items failed to convert are also printed out.
Convert VCF format files¶
VCF (variant call format) is a flexible and extendable line-oriented text format developed by the 1000 Genome Project. It is useful for representing single nucleotide variants, indels, copy number variants, and structural variants. Chromosomes, coordinates, and reference alleles are updated to a new assembly, and all the other fields are not changed.
Typing command without any arguments will print help message:
$ python2.7 CrossMap.py gff
Screen output:
usage:
CrossMap.py vcf input_chain_file input_VCF_file ref_genome_file output_file
Description:
"input_chain_file" and "input_VCF_file" can be regular or compressed (*.gz, *.Z,
*.z, *.bz, *.bz2, *.bzip2) file, local file or URL (http://, https://, ftp://)
pointing to remote file. "ref_genome_file" is genome sequence file of 'target
assembly' in FASTA foramt.
Example:
CrossMap.py vcf hg19ToHg18.over.chain.gz test.hg19.vcf hg18.fa test.hg18.vcf
Example (Convert VCF file from hg19 to hg18):
$ python CrossMap.py vcf hg19ToHg18.over.chain.gz test.hg19.vcf ../database/genome/hg18.fa test.hg18.vcf
@ 2015-07-27 10:14:23: Read chain_file: ../data/hg19ToHg18.over.chain.gz
@ 2013-11-17 20:53:39: Creating index for ../database/genome/hg18.fa
@ 2015-07-27 10:14:50: Total entries: 497
@ 2015-07-27 10:14:50: Failed to map: 0
$ grep -v '#' test.hg19.vcf |head -10
chr1 10933566 . C G . PASS ADP=13;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:7:13:13:0:13:100%:9.6148E-8:0:36:0:0:8:5
chr1 11187893 . T C . PASS ADP=224;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:133:226:224:0:224:100%:3.6518E-134:0:38:0:0:41:183
chr1 11205058 . C T . PASS ADP=625;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:255:643:625:0:625:100%:0E0:0:37:0:0:294:331
chr1 11292753 . A G . PASS ADP=52;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:27:52:52:2:50:96.15%:9.0394E-28:39:38:0:2:0:50
chr1 11318763 . C G . str10 ADP=88;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:51:88:88:0:88:100%:1.7384E-52:0:38:0:0:1:87
chr1 11319587 . A G . PASS ADP=70;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:40:70:70:0:70:100%:1.0659E-41:0:38:0:0:0:70
chr1 16202995 . C T . PASS ADP=463;WT=0;HET=1;HOM=0;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 0/1:1:463:463:458:5:1.08%:3.0913E-2:37:33:188:270:4:1
chr1 27088546 . A T . PASS ADP=124;WT=0;HET=1;HOM=0;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 0/1:21:124:124:65:59:47.58%:1.7915E-22:37:38:59:6:55:4
chr1 27101390 . T C . str10 ADP=267;WT=0;HET=1;HOM=0;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 0/1:1:267:267:262:5:1.87%:3.0665E-2:32:22:85:177:5:0
chr1 34007097 . T C . PASS ADP=10;WT=0;HET=1;HOM=0;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 0/1:1:10:10:6:4:40%:4.3344E-2:34:32:0:6:0:4
$ grep -v '#' test.hg18.vcf |head -10
1 10856153 . C G . PASS ADP=13;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:7:13:13:0:13:100%:9.6148E-8:0:36:0:0:8:5
1 11110480 . T C . PASS ADP=224;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:133:226:224:0:224:100%:3.6518E-134:0:38:0:0:41:183
1 11127645 . C T . PASS ADP=625;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:255:643:625:0:625:100%:0E0:0:37:0:0:294:331
1 11215340 . A G . PASS ADP=52;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:27:52:52:2:50:96.15%:9.0394E-28:39:38:0:2:0:50
1 11241350 . C G . str10 ADP=88;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:51:88:88:0:88:100%:1.7384E-52:0:38:0:0:1:87
1 11242174 . A G . PASS ADP=70;WT=0;HET=0;HOM=1;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 1/1:40:70:70:0:70:100%:1.0659E-41:0:38:0:0:0:70
1 16075582 . C T . PASS ADP=463;WT=0;HET=1;HOM=0;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 0/1:1:463:463:458:5:1.08%:3.0913E-2:37:33:188:270:4:1
1 26961133 . A T . PASS ADP=124;WT=0;HET=1;HOM=0;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 0/1:21:124:124:65:59:47.58%:1.7915E-22:37:38:59:6:55:4
1 26973977 . T C . str10 ADP=267;WT=0;HET=1;HOM=0;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 0/1:1:267:267:262:5:1.87%:3.0665E-2:32:22:85:177:5:0
1 33779684 . T C . PASS ADP=10;WT=0;HET=1;HOM=0;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 0/1:1:10:10:6:4:40%:4.3344E-2:34:32:0:6:0:4
$ grep -v '#' test.hg18.vcf.unmap #coordinates are still based on hg19
chr14 20084444 . G C . PASS ADP=253;WT=0;HET=1;HOM=0;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 0/1:1:253:253:247:5:1.98%:3.0631E-2:38:39:123:124:5:0
chr14 20086290 . T C . PASS ADP=441;WT=0;HET=1;HOM=0;NC=0 GT:GQ:SDP:DP:RD:AD:FREQ:PVAL:RBQ:ABQ:RDF:RDR:ADF:ADR 0/1:4:441:441:427:14:3.17%:5.4963E-5:37:38:236:191:6:8
NOTE:
- Genome coordinates and reference allele will be updated to target assembly.
- Reference genome is genome sequence of target assembly.
- If the reference genome sequence file (../database/genome/hg18.fa) was not indexed, CrossMap will automatically indexed it (only the first time you run CrossMap).
- Output files: output_file and output_file.unmap.
- In the output VCF file, whether the chromosome IDs contain “chr” or not depends on the format of the input VCF file.
Compare to UCSC liftover tool¶
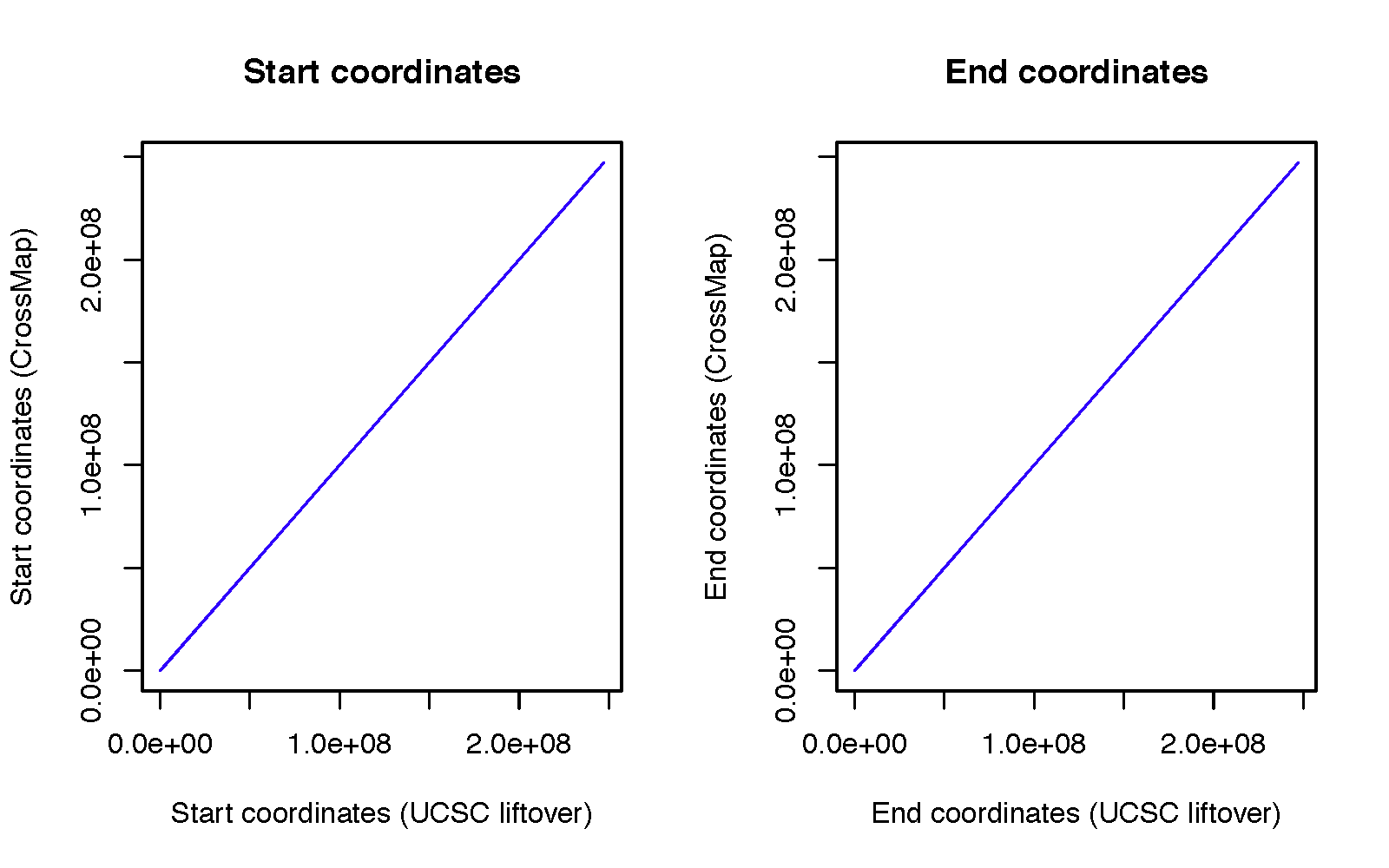
To access the accuracy of CrossMap, we randomly generated 10,000 genome intervals (download from here) with the fixed interval size of 200 bp from hg19. Then we converted them into hg18 using CrossMap and UCSC liftover tool with default configurations. We compare CrossMap to UCSC liftover tool because it is the most widely used tool to convert genome coordinates.
CrossMap failed to convert 613 intervals, and UCSC liftover tool failed to convert 614 intervals. All failed intervals are exactly the same except one region (chr2 90542908 90543108). UCSC failed to convert it because this region needs to be split twice:
| Original (hg19) | Split (hg19) | Target (hg18) |
|---|---|---|
| chr2 90542908 90543108 - | chr2 90542908 90542933 - | chr2 89906445 89906470 - |
| chr2 90542908 90543108 - | chr2 90542933 90543001 - | chr2 87414583 87414651 - |
| chr2 90542908 90543108 - | chr2 90543010 90543108 - | chr2 87414276 87414374 - |
For genome intervals that were successfully converted to hg18, the start and end coordinates are exactly the same between UCSC conversion and CrossMap conversion.
Citation¶
Zhao, H., Sun, Z., Wang, J., Huang, H., Kocher, J.-P., & Wang, L. (2013). CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics (Oxford, England), btt730.
LICENSE¶
CrossMap is distributed under GNU General Public License
This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version. This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details. You should have received a copy of the GNU General Public License along with this program; if not, write to the Free Software Foundation, Inc., 51 Franklin Street, Fifth Floor, Boston, MA 02110-1301 USA


